The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change
Abstract
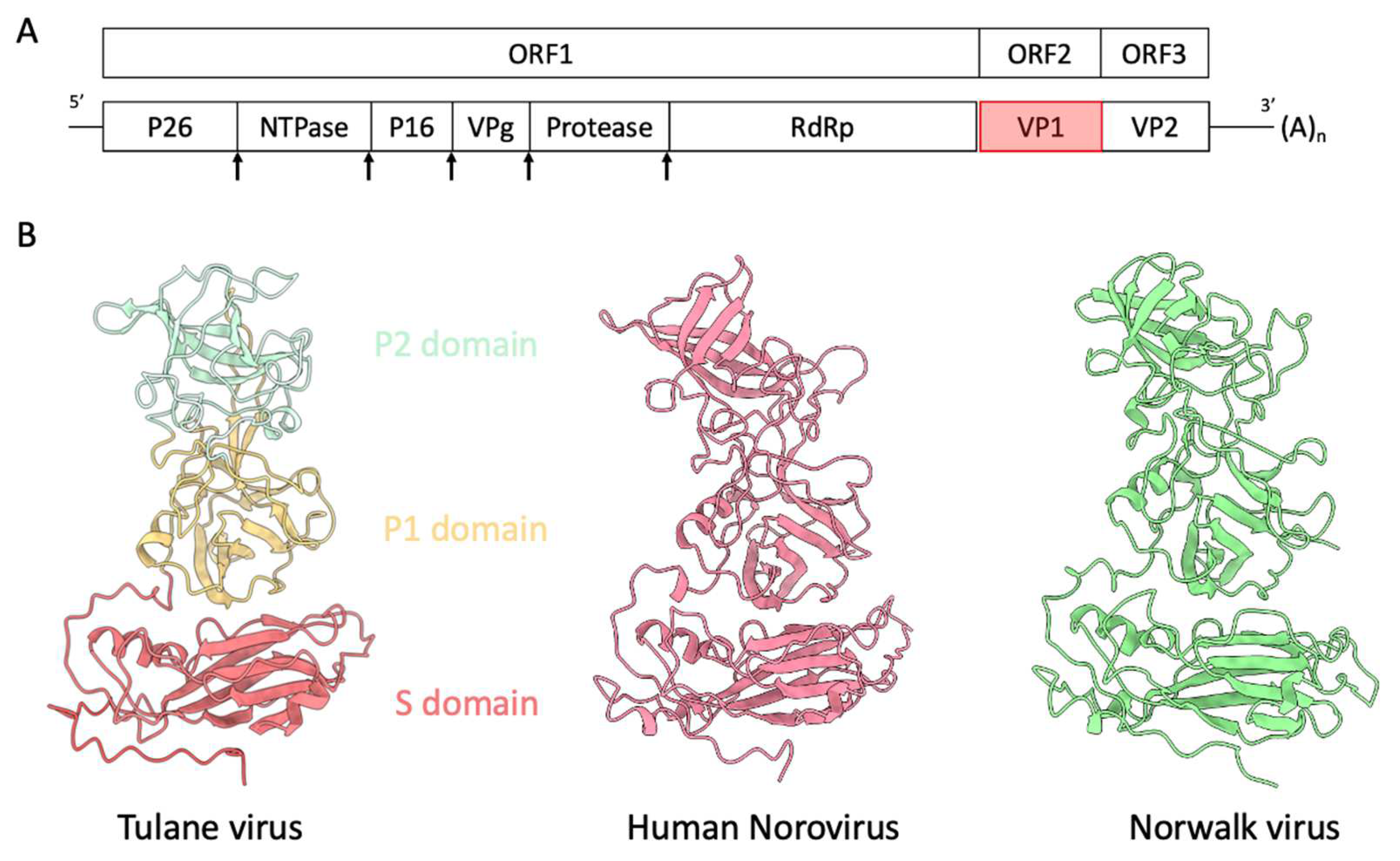
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Purification of Tulane Virus
2.2. Viral RNA Extraction and Genome Sequencing
2.3. Saliva-Based ELISA for Measurement of HBGA Binding
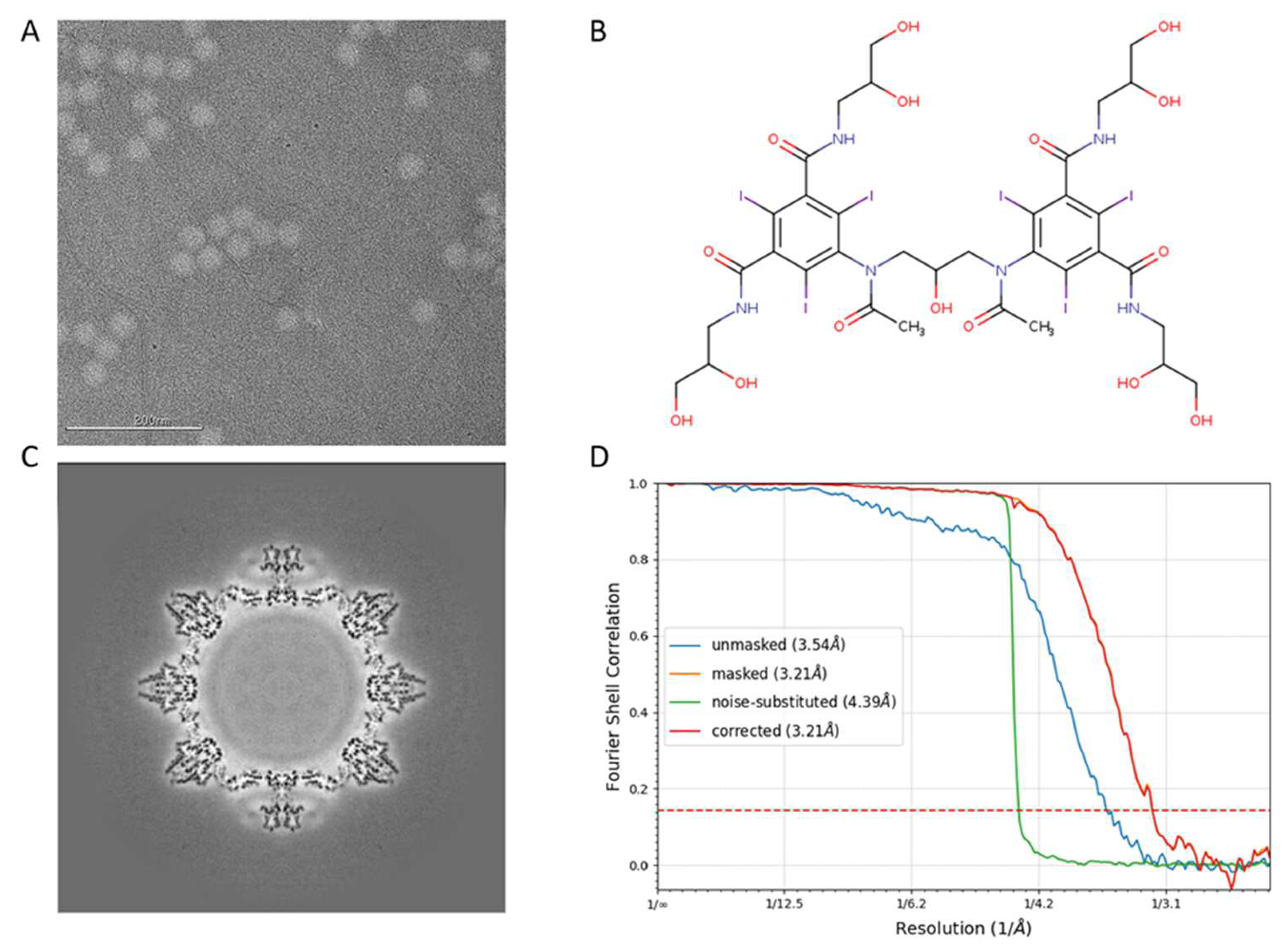
2.4. Cryo-EM Sample Grid Preparation and Data Acquisition
2.5. Image Processing
2.6. Model Refinement
3. Results
3.1. A New TV Variant Has Lost Its Ability to Bind to the Type B HBGA Receptor
3.2. Sequence Analysis of the 9-6-17 TV Strain
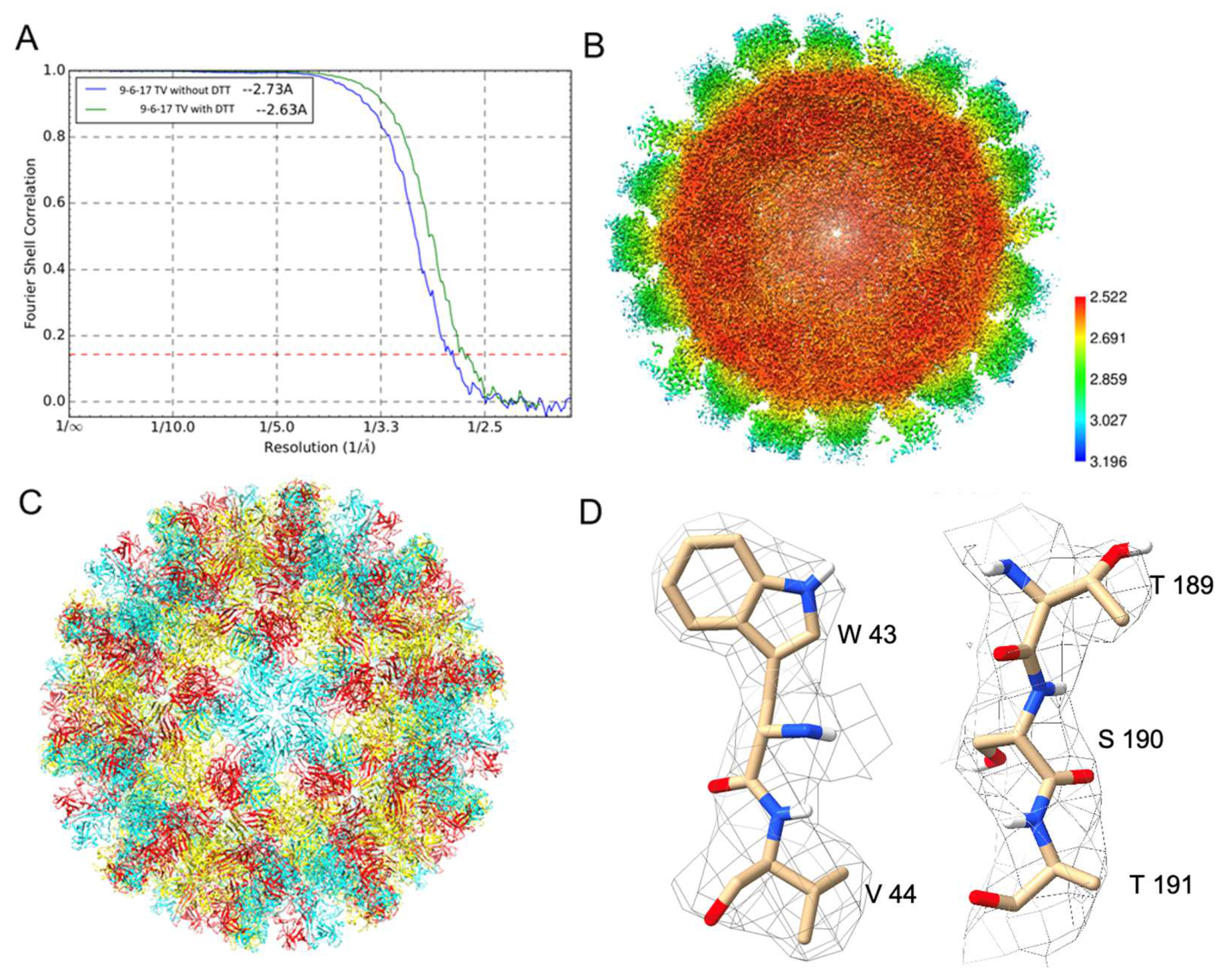
3.3. Structure Determination of the 9-6-17 TV
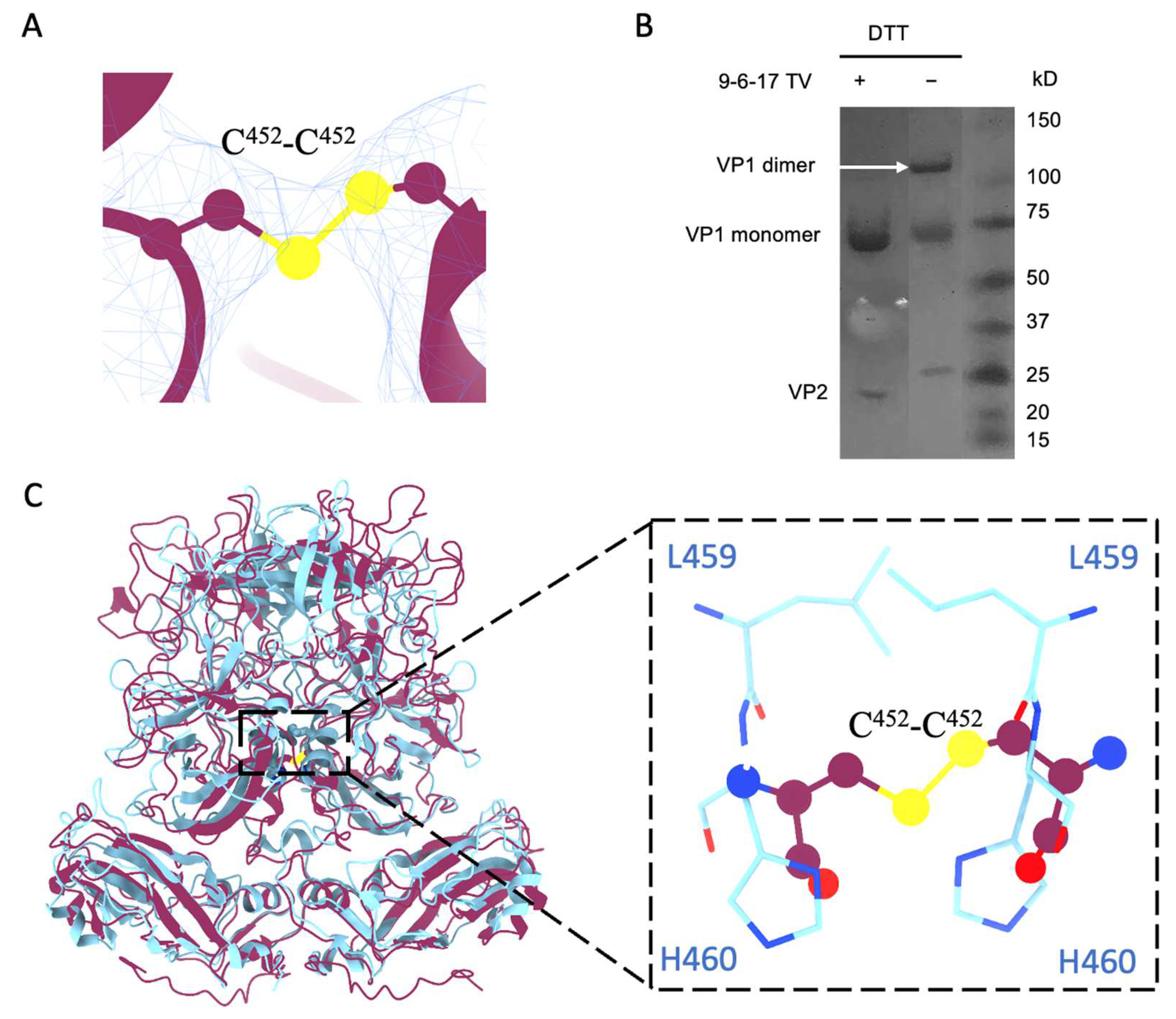
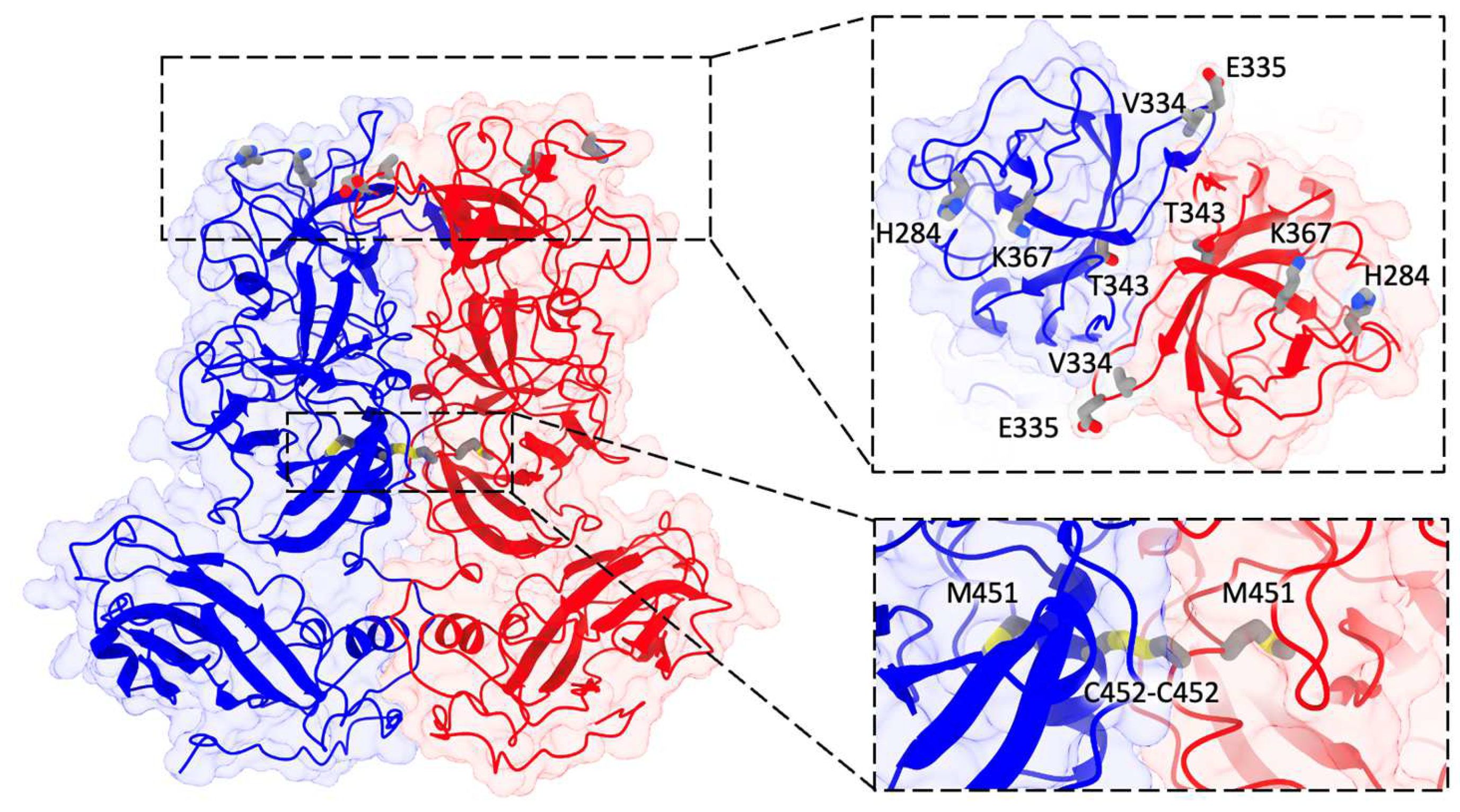
3.4. Disulfide Bond from the R452C Mutation Stabilizes Dimer Interaction
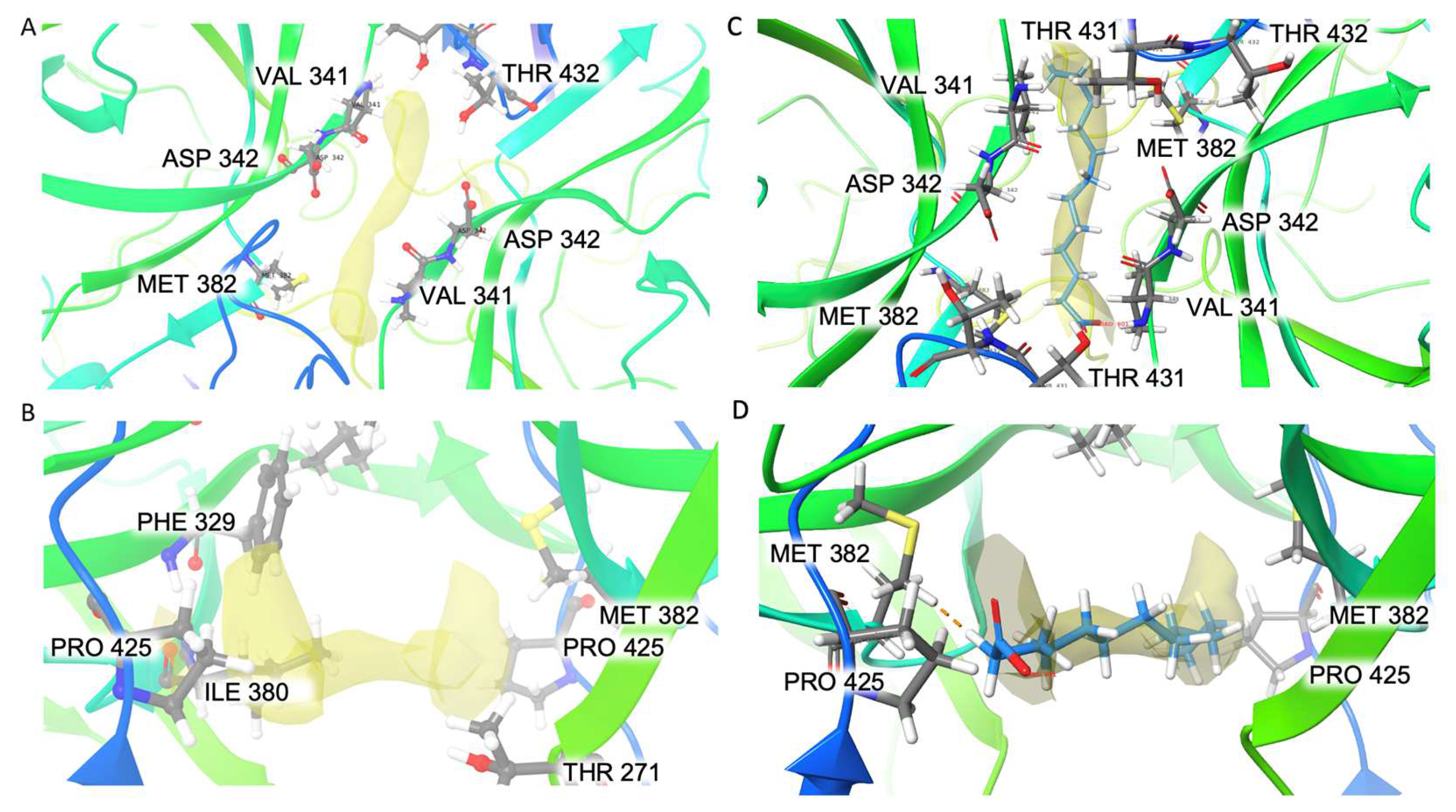
3.5. Elongated Extra Density in the Hydrophobic Pocket in the P Dimer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vinjé, J.; Estes, M.K.; Esteves, P.; Green, K.Y.; Katayama, K.; Knowles, N.J.; L’Homme, Y.; Martella, V.; Vennema, H.; White, P.A.; et al. ICTV Virus Taxonomy Profile: Caliciviridae. J. Gen. Virol. 2019, 100, 1469–1470. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; Tully, D.C.; Mans, J.; Niendorf, S.; Barclay, L.; Cannon, J.L.; Montmayeur, A.M.; Pan, C.Y.; Page, N.; Williams, R.; et al. Emergence of Novel Norovirus GII.4 Variant. Emerg. Infect. Dis. 2024, 30, 163–167. [Google Scholar] [CrossRef]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.L.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed]
- Farkas, T.; Sestak, K.; Wei, C.; Jiang, X. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 2008, 82, 5408–5416. [Google Scholar] [CrossRef]
- Conley, M.J.; McElwee, M.; Azmi, L.; Gabrielsen, M.; Byron, O.; Goodfellow, I.G.; Bhella, D. Calicivirus VP2 forms a portal-like assembly following receptor engagement. Nature 2019, 565, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Wei, C.; Huang, P.; Fan, Q.; Quigley, C.; Xia, M.; Fang, H.; Zhang, X.; Zhong, W.; Klassen, J.S.; et al. Tulane virus recognizes sialic acids as cellular receptors. Sci. Rep. 2015, 5, 11784. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Farkas, T.; Sestak, K.; Jiang, X. Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J. Virol. 2008, 82, 11429–11436. [Google Scholar] [CrossRef] [PubMed]
- Ravn, V.; Dabelsteen, E. Tissue distribution of histo-blood group antigens. APMIS 2000, 108, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Heggelund, J.E.; Varrot, A.; Imberty, A.; Krengel, U. Histo-blood group antigens as mediators of infections. Curr. Opin. Struct. Biol. 2017, 44, 190–200. [Google Scholar] [CrossRef]
- Yu, G.; Li, K.; Jiang, W. Antibody-based affinity cryo-EM grid. Methods 2016, 100, 16–24. [Google Scholar] [CrossRef]
- Yu, G.; Zhang, D.; Guo, F.; Tan, M.; Jiang, X.; Jiang, W. Cryo-EM structure of a novel calicivirus, Tulane virus. PLoS ONE 2013, 8, e59817. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef]
- Grant, T.; Rohou, A.; Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife 2018, 7, e35383. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Guo, F.; Jiang, W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 2014, 1117, 401–443. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Gonzalez, B.; Vago, F.S.; Jiang, W. High resolution single particle Cryo-EM refinement using JSPR. Prog. Biophys. Mol. Biol. 2021, 160, 37–42. [Google Scholar] [CrossRef]
- Zivanov, J.; Oton, J.; Ke, Z.; von Kugelgen, A.; Pyle, E.; Qu, K.; Morado, D.; Castano-Diez, D.; Zanetti, G.; Bharat, T.A.M.; et al. A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Elife 2022, 11, e83724. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Kimanius, D.; Forsberg, B.O.; Scheres, S.H.; Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 2016, 5, e18722. [Google Scholar] [CrossRef]
- Kimanius, D.; Dong, L.; Sharov, G.; Nakane, T.; Scheres, S.H.W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021, 478, 4169–4185. [Google Scholar] [CrossRef]
- Fernandez-Leiro, R.; Scheres, S.H.W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 2017, 73, 496–502. [Google Scholar] [CrossRef]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garcia, R.; Gomez-Blanco, J.; Cuervo, A.; Carazo, J.M.; Sorzano, C.O.S.; Vargas, J. DeepEMhancer: A deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021, 4, 874. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Song, Y.; Barad, B.A.; Cheng, Y.; Fraser, J.S.; DiMaio, F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 2016, 5, e17219. [Google Scholar] [CrossRef] [PubMed]
- Tange, O. GNU Parallel—Line Power Tool—Login. USENIX Mag. 2011, 36, 42–47. [Google Scholar]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, P.; Zou, L.; Lowary, T.L.; Tan, M.; Jiang, X. Tulane virus recognizes the A type 3 and B histo-blood group antigens. J. Virol. 2015, 89, 1419–1427. [Google Scholar] [CrossRef]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Salmen, W.; Chen, R.; Zhou, Y.; Neill, F.; Crowe Jr, J.E.; Prasad, B.V. Atomic structure of the predominant GII.4 human norovirus capsid reveals novel stability and plasticity. Nat. Commun. 2022, 13, 1241. [Google Scholar] [CrossRef]
- Plevka, P.; Perera, R.; Yap, M.L.; Cardosa, J.; Kuhn, R.J.; Rossmann, M.G. Structure of human enterovirus 71 in complex with a capsid-binding inhibitor. Proc. Natl. Acad. Sci. USA 2013, 110, 5463–5467. [Google Scholar] [CrossRef]
- Almand, E.A.; Moore, M.D.; Jaykus, L.A. Norovirus Binding to Ligands Beyond Histo-Blood Group Antigens. Front. Microbiol. 2017, 8, 2549. [Google Scholar] [CrossRef]
- Nelson, C.A.; Wilen, C.B.; Dai, Y.N.; Orchard, R.C.; Kim, A.S.; Stegeman, R.A.; Hsieh, L.L.; Smith, T.J.; Virgin, H.W.; Fremont, D.H. Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. USA 2018, 115, E9201–E9210. [Google Scholar] [CrossRef] [PubMed]



| 9-6-17 TV | |
|---|---|
| Sequence length (bp) | 6701 |
| Number of nucleotide mutations in total | 31 |
| Number of nucleotide mutations in ORF2 | 9 |
| Number of nucleotide mutations in ORF3 | 7 |
| Number of amino acid mutations in total | 18 |
| Number of amino acid mutations in VP1 | 8 |
| Number of amino acid mutations in VP2 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Huang, P.; Xu, X.; Vago, F.S.; Li, K.; Klose, T.; Jiang, X.J.; Jiang, W. The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change. Biomolecules 2024, 14, 119. https://doi.org/10.3390/biom14010119
Sun C, Huang P, Xu X, Vago FS, Li K, Klose T, Jiang XJ, Jiang W. The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change. Biomolecules. 2024; 14(1):119. https://doi.org/10.3390/biom14010119
Chicago/Turabian StyleSun, Chen, Pengwei Huang, Xueyong Xu, Frank S. Vago, Kunpeng Li, Thomas Klose, Xi Jason Jiang, and Wen Jiang. 2024. "The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change" Biomolecules 14, no. 1: 119. https://doi.org/10.3390/biom14010119
APA StyleSun, C., Huang, P., Xu, X., Vago, F. S., Li, K., Klose, T., Jiang, X. J., & Jiang, W. (2024). The 2.6 Å Structure of a Tulane Virus Variant with Minor Mutations Leading to Receptor Change. Biomolecules, 14(1), 119. https://doi.org/10.3390/biom14010119






