A Bacterial Platform for Studying Ubiquitination Cascades Anchored by SCF-Type E3 Ubiquitin Ligases
Abstract
:1. Introduction
2. Materials and Methods
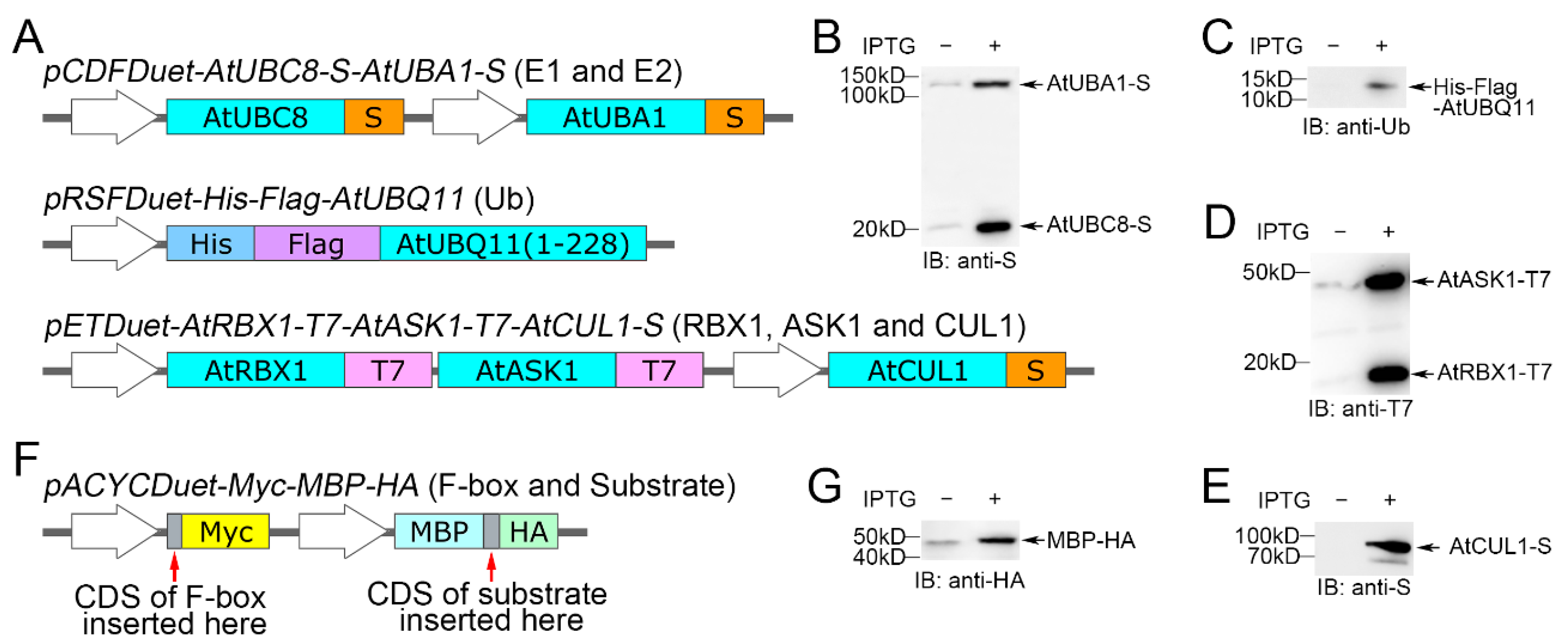
2.1. Plasmid Design for the Expression of Proteins Associated with the Ubiquitination Cascade
2.2. Plasmid Construction
2.3. Co-Expression of Recombinant Proteins in E. coli
2.4. SDS-PAGE and Immunoblot Analysis for Detecting the Ubiquitination Cascade
2.5. Accession Numbers
3. Results
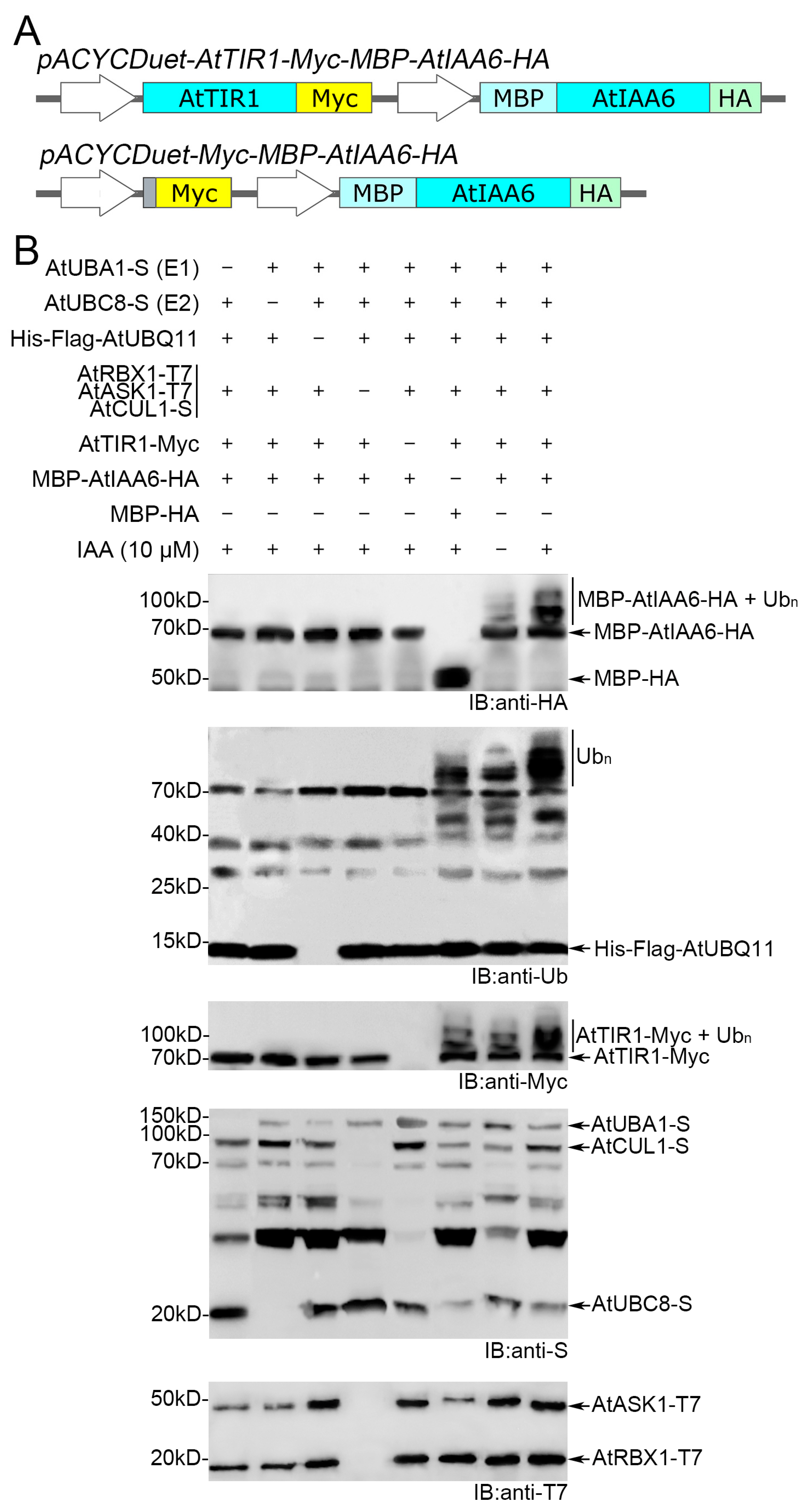
3.1. Auto-Ubiquitination Activity of AtSCFTIR1 in Recombinant E. coli
3.2. AtSCFTIR1 Can Catalyze AtIAA6 Ubiquitination in the Bacterial System
3.3. SCF Complex Composed of Heterologous Subunits Showed Ubiquitin Ligase Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pickart, C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Callis, J. The Ubiquitination Machinery of the Ubiquitin System. Arab. Book 2014, 12, e0174. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Hatfield, P.M.; Gosink, M.M.; Carpenter, T.B.; Vierstra, R.D. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 1997, 11, 213–226. [Google Scholar] [CrossRef]
- Bachmair, A.; Novatchkova, M.; Potuschak, T.; Eisenhaber, F. Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 2001, 6, 463–470. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef]
- Xu, L.; Ménard, R.; Berr, A.; Fuchs, J.; Cognat, V.; Meyer, D.; Shen, W.H. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. SnapShot: Types of Ubiquitin Ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef]
- Schulman, B.A.; Carrano, A.C.; Jeffrey, P.D.; Bowen, Z.; Kinnucan, E.R.E.; Finnin, M.S.; Elledge, S.J.; Harper, J.W.; Pagano, M.; Pavletich, N.P. Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature 2000, 408, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Hotton, S.K.; Callis, J. Regulation of Cullin RING Ligases. Annu. Rev. Plant Biol. 2008, 59, 467–489. [Google Scholar] [CrossRef]
- Frescas, D.; Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer 2008, 8, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Levin-Kravets, O.; Keren-Kaplan, T.; Attali, I.; Sharon, I.; Tanner, N.; Shapira, D.; Rathi, R.; Persaud, A.; Shohat, N.; Shusterman, A.; et al. E. coli-Based Selection and Expression Systems for Discovery, Characterization, and Purification of Ubiquitylated Proteins. Methods Mol. Biol. 2018, 1844, 155–166. [Google Scholar] [CrossRef]
- Keren-Kaplan, T.; Attali, I.; Motamedchaboki, K.; Davis, B.A.; Tanner, N.; Reshef, Y.; Laudon, E.; Kolot, M.; Levin-Kravets, O.; Kleifeld, O.; et al. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. EMBO J. 2012, 31, 378–390. [Google Scholar] [CrossRef]
- Su, L.; Lineberry, N.; Huh, Y.; Soares, L.; Fathman, C.G. A novel E3 ubiquitin ligase substrate screen identifies Rho guanine dissociation inhibitor as a substrate of gene related to anergy in lymphocytes. J. Immunol. 2006, 177, 7559–7566. [Google Scholar] [CrossRef]
- Rosenbaum, J.C.; Fredrickson, E.K.; Oeser, M.L.; Garrett-Engele, C.M.; Locke, M.N.; Richardson, L.A.; Nelson, Z.W.; Hetrick, E.D.; Milac, T.I.; Gottschling, D.E.; et al. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell 2011, 41, 93–106. [Google Scholar] [CrossRef]
- Han, Y.F.; Sun, J.H.; Yang, J.; Tan, Z.Y.; Luo, J.J.; Lu, D.P. Reconstitution of the plant ubiquitination cascade in bacteria using a synthetic biology approach. Plant J. 2017, 91, 766–776. [Google Scholar] [CrossRef]
- Gray, W.M.; del Pozo, J.C.; Walker, L.; Hobbie, L.; Risseeuw, E.; Banks, T.; Crosby, W.L.; Yang, M.; Ma, H.; Estelle, M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999, 13, 1678–1691. [Google Scholar] [CrossRef]
- Cai, L.; Liu, L.; Li, L.; Jia, L. SCFFBXO28-mediated self-ubiquitination of FBXO28 promotes its degradation. Cell. Signal. 2020, 65, 109440. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Friml, J. Fourteen Stations of Auxin. Cold Spring Harb. Perspect. Biol. 2022, 14, a039859. [Google Scholar] [CrossRef]
- Stone, S.L.; HauksdótTir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional Analysis of the RING-Type Ubiquitin Ligase Family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Scaglione, K.M.; Bansal, P.K.; Deffenbaugh, A.E.; Kiss, A.; Moore, J.M.; Korolev, S.; Cocklin, R.; Goebl, M.; Kitagawa, K.; Skowyra, D. SCF E3-Mediated Autoubiquitination Negatively Regulates Activity of Cdc34 E2 but Plays a Nonessential Role in the Catalytic Cycle In Vitro and In Vivo. Mol. Cell. Biol. 2007, 27, 5860–5870. [Google Scholar] [CrossRef]
- Yen, H.-C.S.; Elledge, S.J. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 2008, 322, 923–929. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, Y.; Moss, B.L.; Bargmann, B.O.R.; Wang, R.H.; Prigge, M.; Nemhauser, J.L.; Estelle, M. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat. Plants 2015, 1, 1–8. [Google Scholar] [CrossRef]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D.; et al. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 2017, 8, 15706. [Google Scholar] [CrossRef]
- Niemeyer, M.; Castillo, E.M.; Ihling, C.H.; Iacobucci, C.; Wilde, V.; Hellmuth, A.; Hoehenwarter, W.; Samodelov, S.L.; Zurbriggen, M.D.; Kastritis, P.L.; et al. Flexibility of intrinsically disordered degrons in AUX/IAA proteins reinforces auxin co-receptor assemblies. Nat. Commun. 2020, 11, 2277. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef]
- Skaar, J.R.; D’ANgiolella, V.; Pagan, J.K.; Pagano, M. SnapShot: F Box Proteins II. Cell 2009, 137, 1358.e1–1358.e2. [Google Scholar] [CrossRef]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. SnapShot: F Box Proteins I. Cell 2009, 137, 1160–1160.e1. [Google Scholar] [CrossRef]
- Hua, Z.; Zou, C.; Shiu, S.-H.; Vierstra, R.D. Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 2011, 6, e16219. [Google Scholar] [CrossRef]
- Thomas, J.H. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006, 16, 1017–1030. [Google Scholar] [CrossRef]
- Hua, Z.; Pool, J.E.; Schmitz, R.J.; Schultz, M.D.; Shiu, S.-H.; Ecker, J.R.; Vierstra, R.D. Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 16927–16932. [Google Scholar] [CrossRef]
- Saxena, H.; Negi, H.; Sharma, B. Role of F-box E3-ubiquitin ligases in plant development and stress responses. Plant Cell Rep. 2023, 42, 1133–1146. [Google Scholar] [CrossRef]
- Zhang, X.; Gonzalez-Carranza, Z.H.; Zhang, S.; Miao, Y.; Liu, C.-J.; Roberts, J.A. F-Box Proteins in Plants. Annu. Plant Rev. Online 2019, 2, 307–328. [Google Scholar] [CrossRef]
- Abd-Hamid, N.-A.; Ahmad-Fauzi, M.-I.; Zainal, Z.; Ismail, I. Diverse and dynamic roles of F-box proteins in plant biology. Planta 2020, 251, 68. [Google Scholar] [CrossRef]
- Lee, E.K.; A Diehl, J. SCFs in the new millennium. Oncogene 2014, 33, 2011–2018. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xie, Q.; Hu, C.; Xu, J.; Chen, L.; Li, Y.; Luo, C. F-box proteins and gastric cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Int. J. Med. Sci. 2024, 21, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Turek, I.; Tischer, N.; Lassig, R.; Trujillo, M. Multi-tiered pairing selectivity between E2 ubiquitin-conjugating enzymes and E3 ligases. J. Biol. Chem. 2018, 293, 16324–16336. [Google Scholar] [CrossRef] [PubMed]


| Protein | AtUBC8-S | AtUBA1-S | His-Flag-Ub | AtRBX1-T7 | AtASK1-T7 | AtCUL1-S | MBP-HA |
| MW | 20.9 kD | 123.9 kD | 13.6 kD | 17.9 kD | 44.5 kD | 90.8 kD | 46.0 kD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, Z.-X.; Wang, J.-L.; Li, Y.-Y.; Liang, L.-Y.; Tan, Y.-T.; Wang, Z.-H.; Li, B.-L.; Guo, G.-Q.; Wang, L.; Wu, L. A Bacterial Platform for Studying Ubiquitination Cascades Anchored by SCF-Type E3 Ubiquitin Ligases. Biomolecules 2024, 14, 1209. https://doi.org/10.3390/biom14101209
Pu Z-X, Wang J-L, Li Y-Y, Liang L-Y, Tan Y-T, Wang Z-H, Li B-L, Guo G-Q, Wang L, Wu L. A Bacterial Platform for Studying Ubiquitination Cascades Anchored by SCF-Type E3 Ubiquitin Ligases. Biomolecules. 2024; 14(10):1209. https://doi.org/10.3390/biom14101209
Chicago/Turabian StylePu, Zuo-Xian, Jun-Li Wang, Yu-Yang Li, Luo-Yu Liang, Yi-Ting Tan, Ze-Hui Wang, Bao-Lin Li, Guang-Qin Guo, Li Wang, and Lei Wu. 2024. "A Bacterial Platform for Studying Ubiquitination Cascades Anchored by SCF-Type E3 Ubiquitin Ligases" Biomolecules 14, no. 10: 1209. https://doi.org/10.3390/biom14101209
APA StylePu, Z.-X., Wang, J.-L., Li, Y.-Y., Liang, L.-Y., Tan, Y.-T., Wang, Z.-H., Li, B.-L., Guo, G.-Q., Wang, L., & Wu, L. (2024). A Bacterial Platform for Studying Ubiquitination Cascades Anchored by SCF-Type E3 Ubiquitin Ligases. Biomolecules, 14(10), 1209. https://doi.org/10.3390/biom14101209







