EMC1 Is Required for the Sarcoplasmic Reticulum and Mitochondrial Functions in the Drosophila Muscle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Strains and Genetics
2.2. Generation of UAS-EMC1 Transgenic Fly Lines
2.3. Longevity, Locomotor, and Feeding Assays
2.4. Nucleic Acid Extraction and Quantitative Real-Time PCR
2.5. Preparation of Anti-EMC1 Antibody and Immunoblot Analyses
2.6. Immunostaining of Embryo, Larvae, and Adult Musculature
2.7. Calcium Mobilization Assay and Measurements of Mitochondrial Membrane Potential and Respiration
2.8. Electron Microscopy Analyses
3. Results
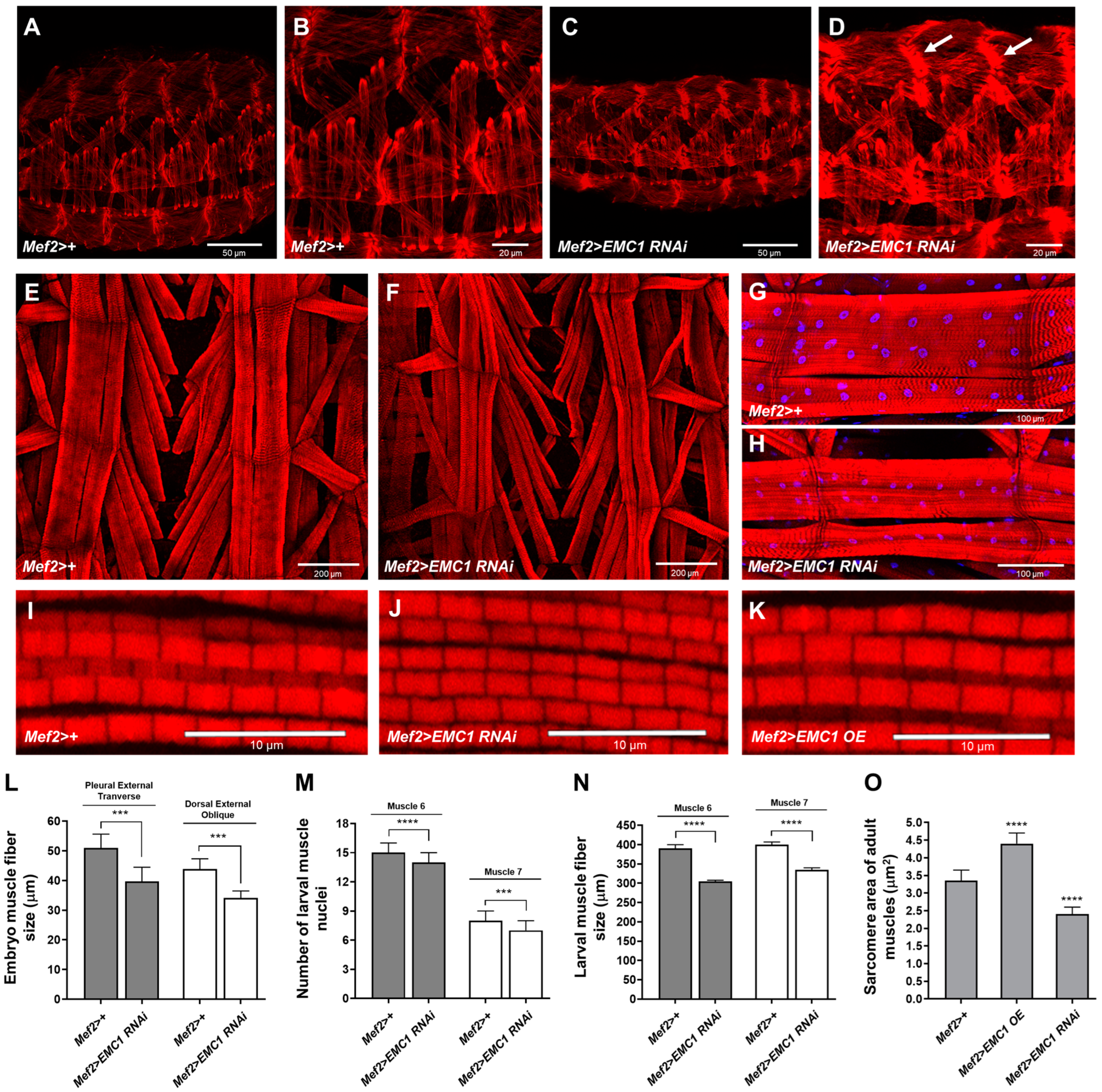
3.1. EMC1 Knockdown in the Drosophila Musculature Triggers Muscle Deformation and Severe Motility Defects
3.2. EMC Is Required for Maintenance of the Sarcoplasmic Reticulum Network and Cytosolic Calcium Homeostasis
3.3. Altered Mitochondrial Shape and Respiration in EMC1-Silenced Fly Muscles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harel, T.; Yesil, G.; Bayram, Y.; Coban-Akdemir, Z.; Charng, W.-L.; Karaca, E.; Al Asmari, A.; Eldomery, M.K.; Hunter, J.V.; Jhangiani, S.N.; et al. Monoallelic and Biallelic Variants in EMC1 Identified in Individuals with Global Developmental Delay, Hypotonia, Scoliosis, and Cerebellar Atrophy. Am. J. Hum. Genet. 2016, 98, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-L.; Rump, P.; Lu, D.; Glassford, M.R.; Mok, J.-W.; Fatih, J.; Basal, A.; Marcogliese, P.C.; Kanca, O.; Rapp, M.; et al. De novo variants in EMC1 lead to neurodevelopmental delay and cerebellar degeneration and affect glial function in Drosophila. Hum. Mol. Genet. 2022, 31, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Geetha, T.S.; Lingappa, L.; Jain, A.R.; Govindan, H.; Mandloi, N.; Murugan, S.; Gupta, R.; Vedam, R. A novel splice variant in EMC1 is associated with cerebellar atrophy, visual impairment, psychomotor retardation with epilepsy. Mol. Genet. Genom. Med. 2017, 6, 282–287. [Google Scholar] [CrossRef]
- Jonikas, M.C.; Collins, S.R.; Denic, V.; Oh, E.; Quan, E.M.; Schmid, V.; Weibezahn, J.; Schwappach, B.; Walter, P.; Weissman, J.S.; et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 2009, 323, 1693–1697. [Google Scholar] [CrossRef]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The ER membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef]
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC Is Required to Initiate Accurate Membrane Protein Topogenesis. Cell 2018, 175, 1507–1519.e16. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, C.J.; Vieira, L.C.; Santos, C.C.; Christianson, J.C.; Jakubec, D.; Strisovsky, K.; Adrain, C.; Domingos, P.M. EMC is required for biogenesis of Xport-A, an essential chaperone of Rhodopsin-1 and the TRP channel. EMBO Rep. 2022, 23, e53210. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Tago, T.; Satoh, T.; Satoh, A.K. ER membrane protein complex is required for the insertions of late-synthesized transmembrane helices of Rh1 in Drosophila photoreceptors. Mol. Biol. Cell 2019, 30, 2890–2900. [Google Scholar] [CrossRef]
- O’Donnell, J.P.; Phillips, B.P.; Yagita, Y.; Juszkiewicz, S.; Wagner, A.; Malinverni, D.; Keenan, R.J.; Miller, E.A.; Hegde, R.S. The architecture of EMC reveals a path for membrane protein insertion. Elife 2020, 9, e57887. [Google Scholar] [CrossRef]
- Satoh, T.; Ohba, A.; Liu, Z.; Inagaki, T.; Satoh, A.K. dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. eLife 2015, 4, e06306. [Google Scholar] [CrossRef]
- Tian, S.; Wu, Q.; Zhou, B.; Choi, M.Y.; Ding, B.; Yang, W.; Dong, M. Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex. Cell Rep. 2019, 28, 2517–2526.e5. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Boulin, T.; Robert, V.J.P.; Richmond, J.E.; Bessereau, J.-L. Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc. Natl. Acad. Sci. USA 2013, 110, E1055–E1063. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Rual, J.-F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A protein complex network of Drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Bircham, P.W.; Maass, D.R.; Roberts, C.A.; Kiew, P.Y.; Low, Y.S.; Yegambaram, M.; Matthews, J.; Jack, C.A.; Atkinson, P.H. Secretory pathway genes assessed by high-throughput microscopy and synthetic genetic array analysis. Mol. Biosyst. 2011, 7, 2589–2598. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhu, X.; Zhang, L. ER membrane complex (EMC): Structure, functions, and roles in diseases. FASEB J. 2024, 38, e23539. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, E.J.; Rutter, G.A. Mitochondrial calcium as a key regulator of mitochondrial ATP production in mammalian cells. Biochim. Biophys. Acta-Bioenerg. 2009, 1787, 1324–1333. [Google Scholar] [CrossRef]
- Xu, J.; Huang, X. Lipid Metabolism at Membrane Contacts: Dynamics and Functions Beyond Lipid Homeostasis. Front. Cell Dev. Biol. 2020, 8, 615856. [Google Scholar] [CrossRef]
- Lahiri, S.; Toulmay, A.; Prinz, W.A. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 2015, 33, 82–87. [Google Scholar] [CrossRef]
- Lahiri, S.; Chao, J.T.; Tavassoli, S.; Wong, A.K.O.; Choudhary, V.; Young, B.P.; Loewen, C.J.R.; Prinz, W.A. A Conserved Endoplasmic Reticulum Membrane Protein Complex (EMC) Facilitates Phospholipid Transfer from the ER to Mitochondria. PLoS Biol. 2014, 12, e1001969. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Rubin, G.M.; Spradling, A.C. Genetic transformation of Drosophila with transposable element vectors. Science 1982, 218, 348–353. [Google Scholar] [CrossRef]
- Steller, H.; Pirrotta, V. P transposons controlled by the heat shock promoter. Mol. Cell. Biol. 1986, 6, 1640–1649. [Google Scholar] [PubMed]
- Benzer, S. Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. USA 1967, 58, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.D.; Becnel, J.; Pandey, U.B. Methods to assay Drosophila behavior. J. Vis. Exp. 2012, e3795. [Google Scholar] [CrossRef]
- Pimenta de Castro, I.; Costa, A.C.; Lam, D.; Tufi, R.; Fedele, V.; Moisoi, N.; Dinsdale, D.; Deas, E.; Loh, S.H.Y.; Martins, L.M. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 2012, 19, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Rera, M.; Bahadorani, S.; Cho, J.; Koehler, C.L.; Ulgherait, M.; Hur, J.H.; Ansari, W.S.; Lo, T.; Jones, D.L.; Walker, D.W. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011, 14, 623–634. [Google Scholar] [CrossRef]
- Dorn, G.W.; Clark, C.F.; Eschenbacher, W.H.; Kang, M.-Y.; Engelhard, J.T.; Warner, S.J.; Matkovich, S.J.; Jowdy, C.C. MARF and Opa1 Control Mitochondrial and Cardiac Function in Drosophila. Circ. Res. 2011, 108, 12–17. [Google Scholar] [CrossRef]
- Liang, C.; Du, F.; Cang, J.; Xue, Z. Pink1 attenuates propofol-induced apoptosis and oxidative stress in developing neurons. J. Anesth. 2018, 32, 62–69. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Wang, K.; Liu, H.; Yang, Z.; Liu, S.; Lu, F. Neuroprotective effects of extract of Acanthopanax senticosus harms on SH-SY5Y cells overexpressing wild-type or A53T mutant α-synuclein. Phytomedicine 2014, 21, 704–711. [Google Scholar] [CrossRef]
- Ponton, F.; Chapuis, M.P.; Pernice, M.; Sword, G.A.; Simpson, S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011, 57, 840–850. [Google Scholar] [CrossRef]
- Narasimha, M.; Brown, N.H. Confocal Microscopy of Drosophilia Embryos. In Cell Biology, 3rd ed.; Celis, J.E., Ed.; Academic Press: Burlington, ON, Canada, 2006; pp. 77–86. [Google Scholar] [CrossRef]
- Brent, J.R.; Werner, K.M.; McCabe, B.D. Drosophila larval NMJ dissection. J. Vis. Exp. 2009, 1107. [Google Scholar] [CrossRef]
- Ramos, R.G.P.; Machado, L.C.H.; Moda, L.M.R. Fluorescent visualization of macromolecules in Drosophila whole mounts. Methods Mol. Biol. 2010, 588, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Christensen, M.E.; Waterhouse, N.J. Measuring Mitochondrial Transmembrane Potential by TMRE Staining. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087361. [Google Scholar] [CrossRef] [PubMed]
- Attrill, H.; Falls, K.; Goodman, J.L.; Millburn, G.H.; Antonazzo, G.; Rey, A.J.; Marygold, S.J. FlyBase: Establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016, 44, D786–D792. [Google Scholar] [CrossRef]
- Pitts, K.R.; Yoon, Y.; Krueger, E.W.; McNiven, M.A. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell 1999, 10, 4403–4417. [Google Scholar] [CrossRef]
- de Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Flis, V.V.; Daum, G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. 2013, 5, a013235. [Google Scholar] [CrossRef]
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 2009, 325, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Naon, D.; Scorrano, L. At the right distance: ER-mitochondria juxtaposition in cell life and death. Biochim. Biophys. Acta 2014, 1843, 2184–2194. [Google Scholar] [CrossRef]
- Rowland, A.A.; Voeltz, G.K. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 2012, 13, 607–625. [Google Scholar] [CrossRef]
- Osman, C.; Voelker, D.R.; Langer, T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011, 192, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, Y.; Schuldiner, M. Staying in touch: The molecular era of organelle contact sites. Trends Biochem. Sci. 2011, 36, 616–623. [Google Scholar] [CrossRef]
- Raturi, A.; Simmen, T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 2013, 1833, 213–224. [Google Scholar] [CrossRef]
- Graveley, B.R.; Brooks, A.N.; Carlson, J.W.; Duff, M.O.; Landolin, J.M.; Yang, L.; Artieri, C.G.; van Baren, M.J.; Boley, N.; Booth, B.W.; et al. The developmental transcriptome of Drosophila melanogaster. Nature 2011, 471, 473–479. [Google Scholar] [CrossRef]
- Hirata, H.; Saint-Amant, L.; Waterbury, J.; Cui, W.; Zhou, W.; Li, Q.; Goldman, D.; Granato, M.; Kuwada, J.Y. Accordion, a zebrafish behavioral mutant, has a muscle relaxation defect due to a mutation in the ATPase Ca2+ pump SERCA1. Development 2004, 131, 5457–5468. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.C.; Betzenhauser, M.J.; Reiken, S.; Meli, A.C.; Umanskaya, A.; Xie, W.; Shiomi, T.; Zalk, R.; Lacampagne, A.; Marks, A.R. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 2011, 14, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chan, D.C. Mitochondrial dynamics--fusion, fission, movement, and mitophagy—In neurodegenerative diseases. Hum. Mol. Genet. 2009, 18, R169–R176. [Google Scholar] [CrossRef]
- van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, a011072. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Y.; Hu, J.; Xiao, J.; Qu, L.; Wang, Z.; Ma, D.; Chen, Y. A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy 2013, 9, 150–163. [Google Scholar] [CrossRef]
- Poole, A.C.; Thomas, R.E.; Andrews, L.A.; McBride, H.M.; Whitworth, A.J.; Pallanck, L.J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. USA 2008, 105, 1638–1643. [Google Scholar] [CrossRef]
- Pesah, Y.; Pham, T.; Burgess, H.; Middlebrooks, B.; Verstreken, P.; Zhou, Y.; Harding, M.; Bellen, H.; Mardon, G. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 2004, 131, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Olzmann, J.A.; Shaler, T.A.; Sowa, M.E.; Bennett, E.J.; Richter, C.M.; Tyler, R.E.; Greenblatt, E.J.; Harper, J.W.; Kopito, R.R. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 2011, 14, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Louie, R.J.; Guo, J.; Rodgers, J.W.; White, R.; Shah, N.; Pagant, S.; Kim, P.; Livstone, M.; Dolinski, K.; McKinney, B.A.; et al. A yeast phenomic model for the gene interaction network modulating CFTR-ΔF508 protein biogenesis. Genome Med. 2012, 4, 103. [Google Scholar] [CrossRef]
- Wójtowicz, I.; Jabłońska, J.; Zmojdzian, M.; Taghli-Lamallem, O.; Renaud, Y.; Junion, G.; Daczewska, M.; Huelsmann, S.; Jagla, K.; Jagla, T. Drosophila small heat shock protein CryAB ensures structural integrity of developing muscles, and proper muscle and heart performance. Development 2015, 142, 994–1005. [Google Scholar] [CrossRef]



| Primer Name | Primer Sequence (5′ 3′) | Reference |
|---|---|---|
| mtDNA | F-CAACCATTCATTCCAGCCTT R-GAAAATTTTAAATGGCCGCA | [25] |
| Dm_ACT79B | F-CCACGCCATCCTTCGTCTA R-GCACAGCTTCTCCTTGATGTC | [25] |
| EMC1 | F-GCAAGGCCGGGGATTTCACA R-ATGTTATCGGACTGGGTCCT | - |
| EMC3 | F-CAGGACGGCCAGGCAATGAT R-CGCCCGTTTCTGAGTCTTGAA | - |
| EMC5 | F-ACAAACTCTTGCTAATTGCCGG R-TCTGCAGGATAATGTCCAATGG | - |
| EMC6 | F-CTGCGCTGCGGGTATTTTGG R-CACTGGGTGCCCGATTTCAC | - |
| EMC8-9 | F-TTTCCACCAGTGCCTGTATG R-TGCGTAGTAGCCTGCGATTA | - |
| IRE1 | F-TGATGGTGTTCTCCACACTG R-TAAATGCTGCCATCTCGAGG | - |
| PERK | F-ATACTTCCATTCCTGGACCG R-AAGAGCTGCTGTTGGCGTTT | - |
| ATF6 | F-AGAGTCTGCTTCCTTATCAC R-TTGCATTTCCAGATTCACGC | - |
| SPARGEL | F-GGATTCACGAATGCTAAATGTGTTCC R-GATGGGTAGGATGCCGCTCAG | [26] |
| MARF | F-GGCGAGGCGTATCTTATGAC R-AGCTTCTCCTGGCACAA | [27] |
| OPA1 | F-CTCTGAGCACCAAGCTAT R-GGCGCAACTTGATGTCTA | [27] |
| DRP1 | F-TCCATCCAATTGCCCCAAAT R-GACGGGTCACAATACCAGTT | - |
| PINK | F-GCTTTCCCCTACCCTCCAC R-GCACTACATTGACCACCGATT | [28] |
| PARKIN | F-AGCCTCCAAGCCTCTAAATG R-CACGGACTCTTTCTTCATCG | [29] |
| CAMKII | F-CCTGTACGCGTTTTTCGGAC; R-CCTCCTGTATACTGTCATGT | - |
| RPL32A | F-ATGCTAAGCTGTCGCACAAATG R-GTTCGATCCGTAACCGATGT | [30] |
| EF1 | F-GCGTGGGTTTGTGATCAGTT R-GATCTTCTCCTTGCCCATCC | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Couto-Lima, C.A.; Machado, M.C.R.; Anhezini, L.; Oliveira, M.T.; Molina, R.A.d.S.; da Silva, R.R.; Lopes, G.S.; Trinca, V.; Colón, D.F.; Peixoto, P.M.; et al. EMC1 Is Required for the Sarcoplasmic Reticulum and Mitochondrial Functions in the Drosophila Muscle. Biomolecules 2024, 14, 1258. https://doi.org/10.3390/biom14101258
Couto-Lima CA, Machado MCR, Anhezini L, Oliveira MT, Molina RAdS, da Silva RR, Lopes GS, Trinca V, Colón DF, Peixoto PM, et al. EMC1 Is Required for the Sarcoplasmic Reticulum and Mitochondrial Functions in the Drosophila Muscle. Biomolecules. 2024; 14(10):1258. https://doi.org/10.3390/biom14101258
Chicago/Turabian StyleCouto-Lima, Carlos Antonio, Maiaro Cabral Rosa Machado, Lucas Anhezini, Marcos Túlio Oliveira, Roberto Augusto da Silva Molina, Rodrigo Ribeiro da Silva, Gabriel Sarti Lopes, Vitor Trinca, David Fernando Colón, Pablo M. Peixoto, and et al. 2024. "EMC1 Is Required for the Sarcoplasmic Reticulum and Mitochondrial Functions in the Drosophila Muscle" Biomolecules 14, no. 10: 1258. https://doi.org/10.3390/biom14101258






