Circular RNAs in Breast Cancer: An Update
Abstract
:1. Introduction
2. Biogenesis of circRNAs
3. Functions of circRNAs
3.1. MiRNA Sponge
3.2. Interacting with Proteins
3.3. Affecting Parental Gene Expression
3.4. Encoding Proteins or Peptides
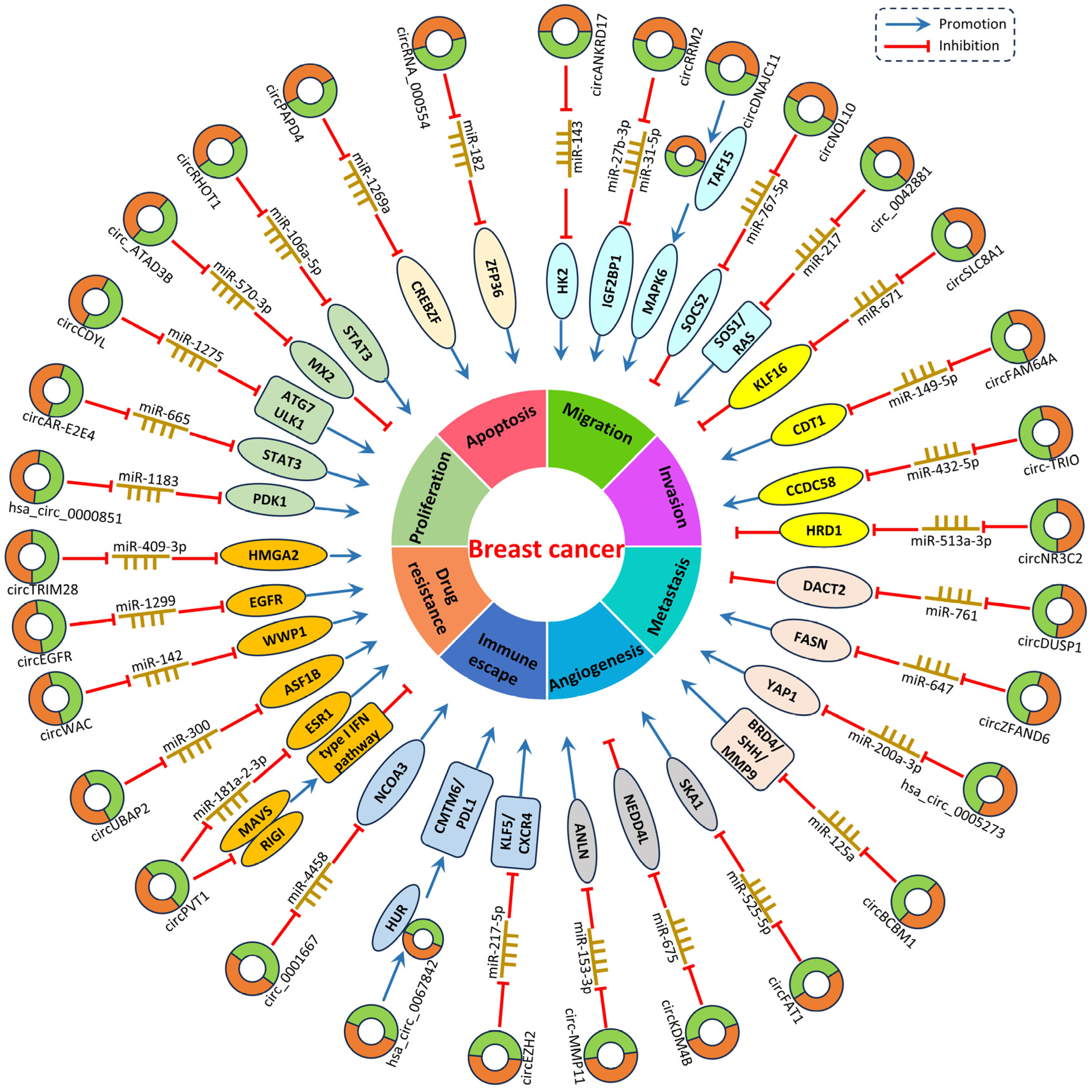
4. CircRNAs in Breast Cancer
4.1. CircRNA Expression Profiles in BC
| CircRNAs | Expression | Target miRNAs | Related Genes and Pathways | Biological Functions | Reference |
|---|---|---|---|---|---|
| circEZH2 | Upregulated | miR-217-5p | KLF5/CXCR4 | Promotes proliferation, invasion, migration, EMT, metastasis | [56] |
| circ_0042881 | Upregulated | miR-217 | SOS1/RAS | Promotes proliferation, invasion, migration, metastasis | [57] |
| circAR-E2E4 | Upregulated | miR-665 | STAT3 | Promotes proliferation | [58] |
| circZFAND6 | Upregulated | miR-647 | FASN | Promotes proliferation, metastasis | [59] |
| hsa_circ_0000851 | Upregulated | miR-1183 | PDK1/p-AKT | Enhances proliferation, migration | [60] |
| hsa_circ_0067842 | Upregulated | / | HuR/CMTM6/PD-L1 | Promotes metastasis, invasion, migration, immune escape | [49] |
| circANKRD17 | Upregulated | miR-143 | HK2 | Promotes growth, invasion, migration, cell cycle progression | [61] |
| circRRM2 | Upregulated | miR-31-5p/miR-27b-3p | IGF2BP1 | Promotes invasion, migration, metastasis | [62] |
| circDNAJC11 | Upregulated | / | TAF15/MAPK6 | Enhances proliferation, migration, invasion, metastasis | [63] |
| circFAM64A | Upregulated | miR-149-5p | CDT1 | Promotes proliferation, invasion, migration, cell cycle progression | [64] |
| circCDYL | Upregulated | miR-1275 | ATG7, ULK1 | Promotes autophagy, proliferation | [65] |
| circRHOT1 | Upregulated | miR-106a-5p | STAT3 | Enhances proliferation, invasion, migration. Inhibits apoptosis, ferroptosis | [66] |
| hsa_circ_0005273 | Upregulated | miR-200a-3p | YAP1, Hippo pathway | Promotes proliferation, migration, metastasis. Regulates cell cycle. | [67] |
| circEGFR | Upregulated | miR-1299 | EGFR | Promotes proliferation, invasion, migration, EMT, THP resistance | [68] |
| circBCBM1 | Upregulated | miR-125a | BRD4/SHH/MMP9 | Enhances proliferation, migration, metastasis | [69] |
| circ-TRIO | Upregulated | miR-432-5p | CCDC58 | Promotes proliferation, invasion, migration, metastasis | [70] |
| circKDM4B | Downregulated | miR-675 | NEDD4L | Suppresses tumor growth, invasion, migration, angiogenesis, metastasis | [71] |
| circNR3C2 | Downregulated | miR-513a-3p | HRD1 | Inhibits proliferation, invasion, migration, metastasis | [72] |
| circ_ATAD3B | Downregulated | miR-570-3p | MX2 | Suppresses proliferation | [73] |
| circNOL10 | Downregulated | miR-767-5p | SOCS2, JAK2/STAT5 | Suppresses proliferation, invasion, migration, EMT | [74] |
| circDUSP1 | Downregulated | miR-761 | DACT2 | Inhibits proliferation, invasion, migration, EMT, metastasis | [75] |
| circPAPD4 | Downregulated | miR-1269a | CREBZF | Suppresses proliferation. Promotes apoptosis | [76] |
| circSLC8A1 | Downregulated | miR-671 | KLF16, PTEN/PI3k/Akt | Suppresses proliferation, migration, invasion | [77] |
| circRNA_000554 | Downregulated | miR-182 | ZFP36 | Induces apoptosis, autophagy. Suppresses EMT, invasion, migration, cell cycle progression. | [78] |
| cSERPINE2 | Upregulated | miR-513a-5p | MALT1/ NF-κB | Enhances proliferation, invasion | [79] |
| circRHOT1 | Upregulated | miR-204-5p | PRMT5 | Promotes proliferation, invasion, migration, EMT | [80] |
| circTBPL1 | Upregulated | miR-653-5p | TPBG | Promotes proliferation, migration, invasion, metastasis | [81] |
| circ_0001142 | Upregulated | miR-361-3p | PIK3CB | Promotes proliferation, metastasis | [82] |
4.2. CircRNAs Regulate the Tumorigenesis and Progression of BC
4.2.1. Tumor Promoters
4.2.2. Tumor Suppressors
5. CircRNAs as Diagnostic and Prognostic Biomarkers in Breast Cancer
5.1. CircRNAs within BC Cells
5.2. Exosomal circRNAs
6. CircRNAs as Therapeutic Targets in Breast Cancer
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Wong, G.L.; Manore, S.G.; Doheny, D.L.; Lo, H.W. STAT family of transcription factors in breast cancer: Pathogenesis and therapeutic opportunities and challenges. Semin. Cancer Biol. 2022, 86, 84–106. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Pellegrino, B.; Hlavata, Z.; Migali, C.; De Silva, P.; Aiello, M.; Willard-Gallo, K.; Musolino, A.; Solinas, C. Luminal Breast Cancer: Risk of Recurrence and Tumor-Associated Immune Suppression. Mol. Diagn. Ther. 2021, 25, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef]
- Derakhshani, A.; Rezaei, Z.; Safarpour, H.; Sabri, M.; Mir, A.; Sanati, M.A.; Vahidian, F.; Gholamiyan Moghadam, A.; Aghadoukht, A.; Hajiasgharzadeh, K.; et al. Overcoming trastuzumab resistance in HER2-positive breast cancer using combination therapy. J. Cell. Physiol. 2020, 235, 3142–3156. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. BCR 2020, 22, 61. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA A Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. TIG 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, T.; Li, T.; Zhao, X. Hsa_circRNA_102002 facilitates metastasis of papillary thyroid cancer through regulating miR-488-3p/HAS2 axis. Cancer Gene Ther. 2021, 28, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zhu, H.; Wang, Z.; Huang, J.; Zhu, Y.; Fan, G.; Wang, Y.; Chen, X.; Zhou, G. Circular RNA circFIRRE drives osteosarcoma progression and metastasis through tumorigenic-angiogenic coupling. Mol. Cancer 2022, 21, 167. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, X.; Qiao, L.; Wang, H. CircRNA circ_0000554 promotes ovarian cancer invasion and proliferation by regulating miR-567. Environ. Sci. Pollut. Res. Int. 2022, 29, 19072–19080. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.; Coca-Prados, M.J.N. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chu, Z.; Lai, W.; Lan, Q.; Zeng, Y.; Lu, D.; Jin, S.; Xu, H.; Su, P.; Yin, D.; et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J. Hematol. Oncol. 2021, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Cardamone, G.; Paraboschi, E.M.; Rimoldi, V.; Duga, S.; Soldà, G.; Asselta, R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 576. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Panda, A.C.; De, S.; Grammatikakis, I.; Munk, R.; Yang, X.; Piao, Y.; Dudekula, D.B.; Abdelmohsen, K.; Gorospe, M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017, 45, e116. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Ferrero, G.; Licheri, N.; Coscujuela Tarrero, L.; De Intinis, C.; Miano, V.; Calogero, R.A.; Cordero, F.; De Bortoli, M.; Beccuti, M. Docker4Circ: A Framework for the Reproducible Characterization of circRNAs from RNA-Seq Data. Int. J. Mol. Sci. 2019, 21, 293. [Google Scholar] [CrossRef]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Xu, Y.P.; Dong, Z.N.; Wang, S.W.; Zheng, Y.M.; Zhang, C.; Zhou, Y.Q.; Zhao, Y.J.; Zhao, Y.; Wang, F.; Peng, R.; et al. circHMGCS1-016 reshapes immune environment by sponging miR-1236-3p to regulate CD73 and GAL-8 expression in intrahepatic cholangiocarcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 290. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Sheng, H.; Zhang, D.S.; Jin, Y.; Zhao, B.T.; Chen, N.; Song, K.; Xu, R.H. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer 2021, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Guo, B.Y.; Wang, H.D.; Lin, G.T.; Lan, T.J.; Ying, H.; Xu, J. CircRNA hsa_circ_0014130 function as a miR-132-3p sponge for playing oncogenic roles in bladder cancer via upregulating KCNJ12 expression. Cell Biol. Toxicol. 2022, 38, 1079–1096. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Che, H.; Che, Y.; Hu, M. CircZNF236 facilitates malignant progression in oral squamous cell carcinoma by sequestering miR-145-5p. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2023, 25, 1690–1701. [Google Scholar] [CrossRef]
- Du, J.; Lan, T.; Liao, H.; Feng, X.; Chen, X.; Liao, W.; Hou, G.; Xu, L.; Feng, Q.; Xie, K.; et al. CircNFIB inhibits tumor growth and metastasis through suppressing MEK1/ERK signaling in intrahepatic cholangiocarcinoma. Mol. Cancer 2022, 21, 18. [Google Scholar] [CrossRef]
- Yang, R.; Chen, H.; Xing, L.; Wang, B.; Hu, M.; Ou, X.; Chen, H.; Deng, Y.; Liu, D.; Jiang, R.; et al. Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10. Mol. Cancer 2022, 21, 88. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Bian, X.; Wu, C.; Hua, J.; Chang, S.; Yu, T.; Li, H.; Li, Y.; Hu, S.; et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2012881118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, J.; Zhang, J.; Shen, H.; Wang, M.; Guo, Z.; Zang, X.; Shi, H.; Gao, J.; Cai, H.; et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol. Cancer 2021, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, S.; He, H.; Ai, K.; Xu, R.; Zhang, L.; Zhu, X. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab. Investig. A J. Technol. Methods Pathol. 2022, 102, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Liu, H.; Wang, Z.; Lu, F.; Chen, W.; Feng, Q.; Miao, Y.; Zhang, J.; Wang, Y.; Chen, Y.; et al. Circular RNA EIF3I promotes papillary thyroid cancer progression by interacting with AUF1 to increase Cyclin D1 production. Oncogene 2023, 42, 3206–3218. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Yang, M.; Wu, C.; Lan, R.; Wang, W.; Li, Y. Circ-SIRT1 inhibits cardiac hypertrophy via activating SIRT1 to promote autophagy. Cell Death Dis. 2021, 12, 1069. [Google Scholar] [CrossRef]
- Ai, N.; Yu, Z.; Xu, X.; Liufu, S.; Wang, K.; Huang, S.; Li, X.; Liu, X.; Chen, B.; Ma, H.; et al. Circular Intronic RNA circTTN Inhibits Host Gene Transcription and Myogenesis by Recruiting PURB Proteins to form Heterotypic Complexes. Int. J. Mol. Sci. 2023, 24, 9859. [Google Scholar] [CrossRef]
- Ma, J.; Du, W.W.; Zeng, K.; Wu, N.; Fang, L.; Lyu, J.; Yee, A.J.; Yang, B.B. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 2754–2768. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Zhu, M.; Dai, Y.; Tong, X.; Zhang, Y.; Zhou, Y.; Cheng, J.; Jiang, Y.; Yang, R.; Wang, X.; Cao, G.; et al. Circ-Udg Derived from Cyprinid Herpesvirus 2 Promotes Viral Replication. Microbiol. Spectr. 2022, 10, e0094322. [Google Scholar] [CrossRef]
- Li, H.; Lan, T.; Liu, H.; Liu, C.; Dai, J.; Xu, L.; Cai, Y.; Hou, G.; Xie, K.; Liao, M.; et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology 2022, 75, 1402–1419. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Su, P.; Liang, Y.; Li, Z.; Zhang, H.; Song, X.; Han, D.; Wang, X.; Liu, Y.; et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Ma, S.; Xu, J.; Ren, X.; Guo, P.; Liu, H.; Li, P.; Yin, F.; Liu, M.; Wang, Q.; et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol. Cancer 2023, 22, 16. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fang, D.; Zhang, C.; Zhao, Z.; Liu, Y.; Zhao, S.; Zhang, N.; Xu, J. Circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction. Mol. Ther. J. Am. Soc. Gene Ther. 2023, 31, 1739–1755. [Google Scholar] [CrossRef]
- Li, Y.; Lei, C.; Xie, Y.; Zhang, J.; Wang, N.; He, W.; Qu, S. Differential expression profile of mRNAs, lncRNAs, and circRNAs reveals potential molecular mechanism in breast cancer. Biosci. Rep. 2022, 42, BSR20220645. [Google Scholar] [CrossRef]
- Li, J.; Dong, X.; Kong, X.; Wang, Y.; Li, Y.; Tong, Y.; Zhao, W.; Duan, W.; Li, P.; Wang, Y.; et al. Circular RNA hsa_circ_0067842 facilitates tumor metastasis and immune escape in breast cancer through HuR/CMTM6/PD-L1 axis. Biol. Direct 2023, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Lyu, M.; Yang, S.; Zhang, J.; Yu, D. CircRNA expression profiles of breast cancer and construction of a circRNA-miRNA-mRNA network. Sci. Rep. 2022, 12, 17765. [Google Scholar] [CrossRef]
- Zheng, M.; Cai, W.H.; Wang, M.F.; Deng, Y.J.; Huang, L.L.; Cao, Y.J. Microarray Profile of Circular RNAs Identifies hsa_circ_0001583 as A New Circular RNA Biomarker for Breast Cancer: A Retrospective Study. Cell J. 2022, 24, 500–505. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Z.; Hu, G.; Zhang, Y.; Feng, Y.; Jiang, Y.; Wang, J. Profiling and integrated analysis of differentially expressed circRNAs as novel biomarkers for breast cancer. J. Cell. Physiol. 2020, 235, 7945–7959. [Google Scholar] [CrossRef]
- Yuan, C.; Zhou, L.; Zhang, L.; Yin, K.; Peng, J.; Sha, R.; Zhang, S.; Xu, Y.; Sheng, X.; Wang, Y.; et al. Identification and integrated analysis of key differentially expressed circular RNAs in ER-positive subtype breast cancer. Epigenomics 2019, 11, 297–321. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Luo, X.; Zhan, X.; Zeng, H.; Duan, S. EMT related circular RNA expression profiles identify circSCYL2 as a novel molecule in breast tumor metastasis. Int. J. Mol. Med. 2020, 45, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Zhang, A.; Li, M.; Pan, L.; Tang, W.; An, M.; Liu, W.; Zhang, J. Circular RNA profile of breast cancer brain metastasis: Identification of potential biomarkers and therapeutic targets. Epigenomics 2018, 10, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Z.; Ou, X.; Wu, P.; Zhang, Y.; Wu, S.; Xiao, X.; Li, Y.; Ye, F.; Tang, H. The FUS/circEZH2/KLF5/ feedback loop contributes to CXCR4-induced liver metastasis of breast cancer by enhancing epithelial-mesenchymal transition. Mol. Cancer 2022, 21, 198. [Google Scholar] [CrossRef]
- Ju, C.; Zhou, M.; Du, D.; Wang, C.; Yao, J.; Li, H.; Luo, Y.; He, F.; He, J. EIF4A3-mediated circ_0042881 activates the RAS pathway via miR-217/SOS1 axis to facilitate breast cancer progression. Cell Death Dis. 2023, 14, 559. [Google Scholar] [CrossRef]
- Xu, H.; Fang, M.; Zuo, B.; Yao, W.; Ren, J.; Zhang, Y. circAR-E2E4-miR-665-STAT3 axis is a potential regulatory network in triple-negative breast cancer. Heliyon 2023, 9, e12654. [Google Scholar] [CrossRef]
- Huang, X.; Tan, W.; Liu, Z.; Fu, X.; Li, Z.; Lai, S.; Li, Q.; Zhong, X.; Qu, F.; Zhang, H.; et al. EIF4A3-induced circZFAND6 promotes breast cancer proliferation and metastasis through the miR-647/FASN axis. Life Sci. 2023, 324, 121745. [Google Scholar] [CrossRef]
- Ji, C.; Zhu, L.; Fang, L. has_circ_0000851 promotes PDK1/p-AKT-mediated cell proliferation and migration by regulating miR-1183 in triple-negative breast cancer. Cell. Signal. 2023, 101, 110494. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, L.F.; Zhang, L.; Miao, Y.; Xi, Y.; Liu, M.F.; Zhang, M.; Li, B. CircANKRD17 promotes glycolysis by inhibiting miR-143 in breast cancer cells. J. Cell. Physiol. 2023, 23, 2765–2777. [Google Scholar] [CrossRef]
- Hao, R.; Zhang, L.; Si, Y.; Zhang, P.; Wang, Y.; Li, B.; Hu, J.; Qi, Y. A novel feedback regulated loop of circRRM2-IGF2BP1-MYC promotes breast cancer metastasis. Cancer Cell Int. 2023, 23, 54. [Google Scholar] [CrossRef]
- Wang, B.; Chen, H.; Deng, Y.; Chen, H.; Xing, L.; Guo, Y.; Wang, M.; Chen, J. CircDNAJC11 interacts with TAF15 to promote breast cancer progression via enhancing MAPK6 expression and activating the MAPK signaling pathway. J. Transl. Med. 2023, 21, 186. [Google Scholar] [CrossRef]
- Maimaiti, Y.; Zhang, N.; Zhang, Y.; Zhou, J.; Song, H.; Wang, S. CircFAM64A enhances cellular processes in triple-negative breast cancer by targeting the miR-149-5p/CDT1 axis. Environ. Toxicol. 2022, 37, 1081–1092. [Google Scholar] [CrossRef]
- Liang, G.; Ling, Y.; Mehrpour, M.; Saw, P.E.; Liu, Z.; Tan, W.; Tian, Z.; Zhong, W.; Lin, W.; Luo, Q.; et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol. Cancer 2020, 19, 65. [Google Scholar] [CrossRef]
- Zhang, H.; Ge, Z.; Wang, Z.; Gao, Y.; Wang, Y.; Qu, X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging 2021, 13, 8115–8126. [Google Scholar] [CrossRef]
- Wang, X.; Ji, C.; Hu, J.; Deng, X.; Zheng, W.; Yu, Y.; Hua, K.; Zhou, X.; Fang, L. Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-hippo signaling pathway. J. Exp. Clin. Cancer Res. 2021, 40, 29. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Chen, C.; Fan, Z.; Zhang, Y.; Ji, J.; Wei, D.; Zhang, F.; Sun, B.; Huang, P.; Ren, L. CircEGFR reduces the sensitivity of pirarubicin and regulates the malignant progression of triple-negative breast cancer via the miR-1299/EGFR axis. Int. J. Biol. Macromol. 2023, 244, 125295. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Liu, W.; Zhu, C.; Li, P.; Wang, L.; Pan, L.; Li, K.; Cai, P.; Meng, M.; Wang, Y.; et al. Circular RNA circBCBM1 promotes breast cancer brain metastasis by modulating miR-125a/BRD4 axis. Int. J. Biol. Sci. 2021, 17, 3104–3117. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Yang, J.; Liang, Y.; Wang, X.; Zhang, N.; Kong, X.; Chen, B.; Wang, L.; Zhao, W.; et al. Circ-TRIO promotes TNBC progression by regulating the miR-432-5p/CCDC58 axis. Cell Death Dis. 2022, 13, 776. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Liu, T.T.; Zhu, W.J.; Liu, H.T.; Zhang, G.H.; Song, L.; Zhao, R.N.; Chen, X.; Gao, P. CircKDM4B suppresses breast cancer progression via the miR-675/NEDD4L axis. Oncogene 2022, 41, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, J.; Jin, W.; Sun, Y.; Xu, Y.; Wang, Y.; Liang, X.; Su, D. CircNR3C2 promotes HRD1-mediated tumor-suppressive effect via sponging miR-513a-3p in triple-negative breast cancer. Mol. Cancer 2021, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Xu, C.; Zhang, Y.; Shan, Y. Circ_ATAD3B inhibits cell proliferation of breast cancer via mediating the miR-570-3p/MX2 axis. Prev. Med. 2023, 173, 107568. [Google Scholar] [CrossRef]
- Wang, F.; Wang, X.; Li, J.; Lv, P.; Han, M.; Li, L.; Chen, Z.; Dong, L.; Wang, N.; Gu, Y. CircNOL10 suppresses breast cancer progression by sponging miR-767-5p to regulate SOCS2/JAK/STAT signaling. J. Biomed. Sci. 2021, 28, 4. [Google Scholar] [CrossRef]
- Huang, S.; Xie, J.; Lei, S.; Fan, P.; Zhang, C.; Huang, Z. CircDUSP1 regulates tumor growth, metastasis, and paclitaxel sensitivity in triple-negative breast cancer by targeting miR-761/DACT2 signaling axis. Mol. Carcinog. 2023, 62, 450–463. [Google Scholar] [CrossRef]
- Zhou, B.; Xue, J.; Wu, R.; Meng, H.; Li, R.; Mo, Z.; Zhai, H.; Chen, X.; Liu, R.; Lai, G.; et al. CREBZF mRNA nanoparticles suppress breast cancer progression through a positive feedback loop boosted by circPAPD4. J. Exp. Clin. Cancer Res. 2023, 42, 138. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, X.; Zai, H.Y.; Jiang, W.; Zhang, K.J.; He, Y.Q.; Hu, Y. circSLC8A1 sponges miR-671 to regulate breast cancer tumorigenesis via PTEN/PI3k/Akt pathway. Genomics 2021, 113, 398–410. [Google Scholar] [CrossRef]
- Mao, Y.; Lv, M.; Cao, W.; Liu, X.; Cui, J.; Wang, Y.; Wang, Y.; Nie, G.; Liu, X.; Wang, H. Circular RNA 000554 represses epithelial-mesenchymal transition in breast cancer by regulating microRNA-182/ZFP36 axis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 11405–11420. [Google Scholar] [CrossRef]
- Zhou, B.; Mo, Z.; Lai, G.; Chen, X.; Li, R.; Wu, R.; Zhu, J.; Zheng, F. Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF-. J. Exp. Clin. Cancer Res. 2023, 42, 48. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, Y.; Ou, J.; Li, Y.; Zhu, N. Exosomal circRNA RHOT1 promotes breast cancer progression by targeting miR-204-5p/ PRMT5 axis. Cancer Cell Int. 2023, 23, 260. [Google Scholar] [CrossRef]
- Ye, F.; Liang, Y.; Wang, Y.; Le Yang, R.; Luo, D.; Li, Y.; Jin, Y.; Han, D.; Chen, B.; Zhao, W.; et al. Cancer-associated fibroblasts facilitate breast cancer progression through exosomal circTBPL1-mediated intercellular communication. Cell Death Dis. 2023, 14, 471. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Shi, W.; Hu, W.; Zhao, Y.; Zhao, X.; Dong, F.; Xin, Y.; Peng, T.; Liu, C. Endoplasmic reticulum stress promotes breast cancer cells to release exosomes circ_0001142 and induces M2 polarization of macrophages to regulate tumor progression. Pharmacol. Res. 2022, 177, 106098. [Google Scholar] [CrossRef]
- Yang, Q.; Li, F.; He, A.T.; Yang, B.B. Circular RNAs: Expression, localization, and therapeutic potentials. Mol. Ther. J. Am. Soc. Gene Ther. 2021, 29, 1683–1702. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Ben Fraj, S.; Naserian, S.; Lorenzini, B.; Goulinet, S.; Mauduit, P.; Uzan, G.; Haouas, H. Human Umbilical Cord Blood Endothelial Progenitor Cell-Derived Extracellular Vesicles Control Important Endothelial Cell Functions. Int. J. Mol. Sci. 2023, 24, 9866. [Google Scholar] [CrossRef] [PubMed]
- Bai, K.; Lee, C.L.; Liu, X.; Li, J.; Cao, D.; Zhang, L.; Hu, D.; Li, H.; Hou, Y.; Xu, Y.; et al. Human placental exosomes induce maternal systemic immune tolerance by reprogramming circulating monocytes. J. Nanobiotechnol. 2022, 20, 86. [Google Scholar] [CrossRef]
- Farhadi, S.; Mohammadi-Yeganeh, S.; Kiani, J.; Hashemi, S.M.; Koochaki, A.; Sharifi, K.; Ghanbarian, H. Exosomal delivery of 7SK long non-coding RNA suppresses viability, proliferation, aggressiveness and tumorigenicity in triple negative breast cancer cells. Life Sci. 2023, 322, 121646. [Google Scholar] [CrossRef]
- Nolan, E.; Lindeman, G.J.; Visvader, J.E. Deciphering breast cancer: From biology to the clinic. Cell 2023, 186, 1708–1728. [Google Scholar] [CrossRef]
- Ling, Y.; Liang, G.; Lin, Q.; Fang, X.; Luo, Q.; Cen, Y.; Mehrpour, M.; Hamai, A.; Liu, Z.; Shi, Y.; et al. circCDYL2 promotes trastuzumab resistance via sustaining HER2 downstream signaling in breast cancer. Mol. Cancer 2022, 21, 8. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Li, Q.; Li, X.; Feng, X. A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ. Toxicol. 2022, 37, 1597–1607. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Li, Q.; Li, X.; Feng, X.; Zeng, K. The novel β-TrCP protein isoform hidden in circular RNA confers trastuzumab resistance in HER2-positive breast cancer. Redox Biol. 2023, 67, 102896. [Google Scholar] [CrossRef]
- Yao, Y.; Li, X.; Cheng, L.; Wu, X.; Wu, B. Circular RNA FAT atypical cadherin 1 (circFAT1)/microRNA-525-5p/spindle and kinetochore-associated complex subunit 1 (SKA1) axis regulates oxaliplatin resistance in breast cancer by activating the notch and Wnt signaling pathway. Bioengineered 2021, 12, 4032–4043. [Google Scholar] [CrossRef]
- Yi, J.; Wang, L.; Hu, G.S.; Zhang, Y.Y.; Du, J.; Ding, J.C.; Ji, X.; Shen, H.F.; Huang, H.H.; Ye, F.; et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. EMBO J. 2023, 42, e112408. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Han, D.; Wang, X.; Li, C.; Chen, T.; Li, W.; Liang, Y.; Luo, D.; Chen, B.; et al. circRNA-SFMBT2 orchestrates ERα activation to drive tamoxifen resistance in breast cancer cells. Cell Death Dis. 2023, 14, 482. [Google Scholar] [CrossRef]
- Yang, S.; Zou, C.; Li, Y.; Yang, X.; Liu, W.; Zhang, G.; Lu, N. Knockdown circTRIM28 enhances tamoxifen sensitivity via the miR-409-3p/HMGA2 axis in breast cancer. Reprod. Biol. Endocrinol. RBE 2022, 20, 146. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Jiang, L.; Lu, L.; Dai, T.; Li, A.; Chen, Y.; Zhang, L. CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. Mol. Cancer 2021, 20, 43. [Google Scholar] [CrossRef]
- Wu, X.; Ren, Y.; Yao, R.; Zhou, L.; Fan, R. Circular RNA circ-MMP11 Contributes to Lapatinib Resistance of Breast Cancer Cells by Regulating the miR-153-3p/ANLN Axis. Front. Oncol. 2021, 11, 639961. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, X.; Zhou, F.; Sun, X.; Li, S. Circular RNA UBAP2 facilitates the cisplatin resistance of triple-negative breast cancer via microRNA-300/anti-silencing function 1B histone chaperone/PI3K/AKT/mTOR axis. Bioengineered 2022, 13, 7197–7208. [Google Scholar] [CrossRef]
- Cui, Y.; Fan, J.; Shi, W.; Zhou, Z. Circ_0001667 knockdown blocks cancer progression and attenuates adriamycin resistance by depleting NCOA3 via releasing miR-4458 in breast cancer. Drug Dev. Res. 2022, 83, 75–87. [Google Scholar] [CrossRef] [PubMed]

| Experimental Model | Method | Differentially Expressed circRNAs | Reference |
|---|---|---|---|
| BC tissues | RNA sequencing | 70 circRNAs were upregulated and 78 circRNAs were downregulated. | [48] |
| BC tissues | CircRNA microarray analysis | 89 downregulated and 129 upregulated circRNAs were reported. | [49] |
| BC tissues | CircRNA high-throughput sequencing | 136 increased circRNAs were found. | [50] |
| BC tissues | CircRNA microarray analysis | 256 upregulated circRNAs and 277 downregulated circRNAs were identified. | [51] |
| BC tissues | ceRNA microarray probes | 2375 and 1995 circRNAs were increased and decreased, respectively. | [52] |
| ER-positive BC tissues | CircRNA microarray analysis | 1700 circRNAs were downregulated and 1953 circRNAs were upregulated. | [53] |
| BC cell lines | High-throughput RNA sequencing | 16 downregulated and 7 upregulated circRNAs were discovered. | [54] |
| BCBM cell lines | RNA-sequencing | 191 circRNAs were decreased and 215 circRNAs were elevated. | [55] |
| Therapeutic Drugs | CircRNAs | Expression | Effect on Drug Resistance | Pathways | Reference |
|---|---|---|---|---|---|
| Pirarubicin | circEGFR | Up | Promoting | circEGFR/miR-1299/EGFR | [68] |
| Trastuzumab | circCDYL2 | Up | Promoting | circCDYL2/GRB7/FAK/AKT and ERK1/2 | [90] |
| Trastuzumab | circ-BGN | Up | Promoting | circBGN/OTUB1/SLC7A11 | [91] |
| Trastuzumab | circ-β-TrCP | Up | Promoting | circ-β-TrCP/β-TrCP-343aa/NRF2 | [92] |
| Oxaliplatin | circFAT1 | Up | Promoting | circFAT1/miR-525-5p/SKA1/Notch and Wnt pathway | [93] |
| Tamoxifen | circPVT1 | Up | Promoting | circPVT1/miR-181a-2-3p/ESR1; circPVT1/MAVS | [94] |
| Tamoxifen | circRNA-SFMBT2 | Up | Promoting | circRNA-SFMBT2/RNF181 | [95] |
| Tamoxifen | circTRIM28 | Up | Promoting | circTRIM28/miR-409-3p/HMGA2 | [96] |
| Paclitaxel | circWAC | Up | Promoting | circWAC/miR-142/WWP1 | [97] |
| Lapatinib | circ-MMP11 | Up | Promoting | circ-MMP11/miR-153-3p/ANLN | [98] |
| Cisplatin | circUBAP2 | Up | Promoting | circUBAP2/miR-300/ASF1B/PI3K/AKT/mTOR | [99] |
| Adriamycin | circ_0001667 | Up | Promoting | circ_0001667/miR-4458/NCOA3 | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, H.; Li, J.; Zhao, Q.; Yang, Q.; Xu, Y. Circular RNAs in Breast Cancer: An Update. Biomolecules 2024, 14, 158. https://doi.org/10.3390/biom14020158
Bao H, Li J, Zhao Q, Yang Q, Xu Y. Circular RNAs in Breast Cancer: An Update. Biomolecules. 2024; 14(2):158. https://doi.org/10.3390/biom14020158
Chicago/Turabian StyleBao, Haolin, Jiehan Li, Qihang Zhao, Qingling Yang, and Yi Xu. 2024. "Circular RNAs in Breast Cancer: An Update" Biomolecules 14, no. 2: 158. https://doi.org/10.3390/biom14020158
APA StyleBao, H., Li, J., Zhao, Q., Yang, Q., & Xu, Y. (2024). Circular RNAs in Breast Cancer: An Update. Biomolecules, 14(2), 158. https://doi.org/10.3390/biom14020158






