Depletion of PHLDB2 Suppresses Epithelial–Mesenchymal Transition and Enhances Anti-Tumor Immunity in Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Real-Time Quantity PCR (RT-qPCR)
2.3. Western Blot
2.4. Immunohistology (IHC)
2.5. sgRNA Transfection
2.6. CCK-8 Assay
2.7. Survival Analysis
2.8. Co-Expression Gene and Survival-Related Gene Analysis
2.9. T Cell Cytotoxicity Assays
2.10. C3H Syngeneic Model
2.11. Statistical Analysis
3. Results
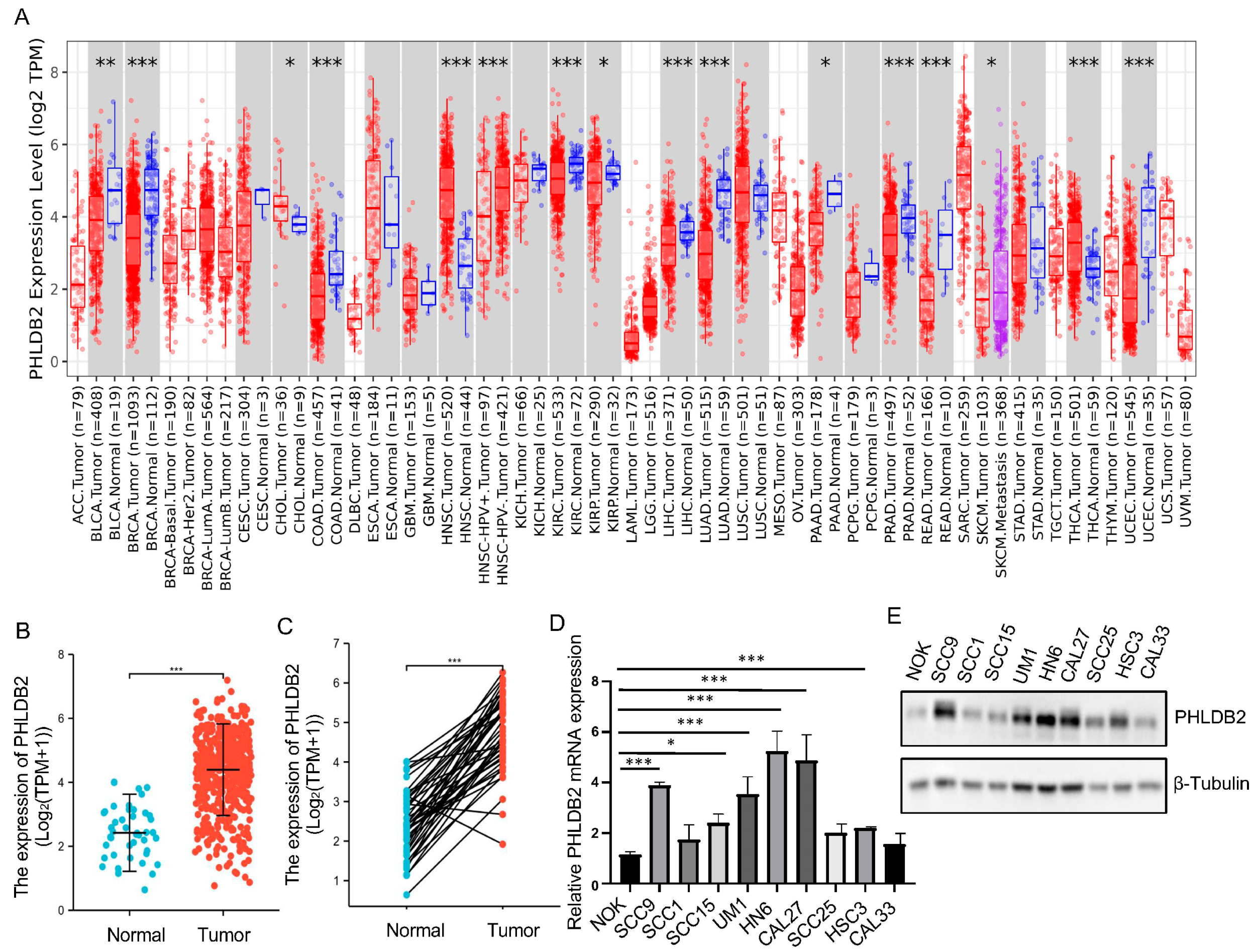
3.1. Higher Expression of PHLDB2 Is Correlated with Poorer Prognosis for HNSCC Patients
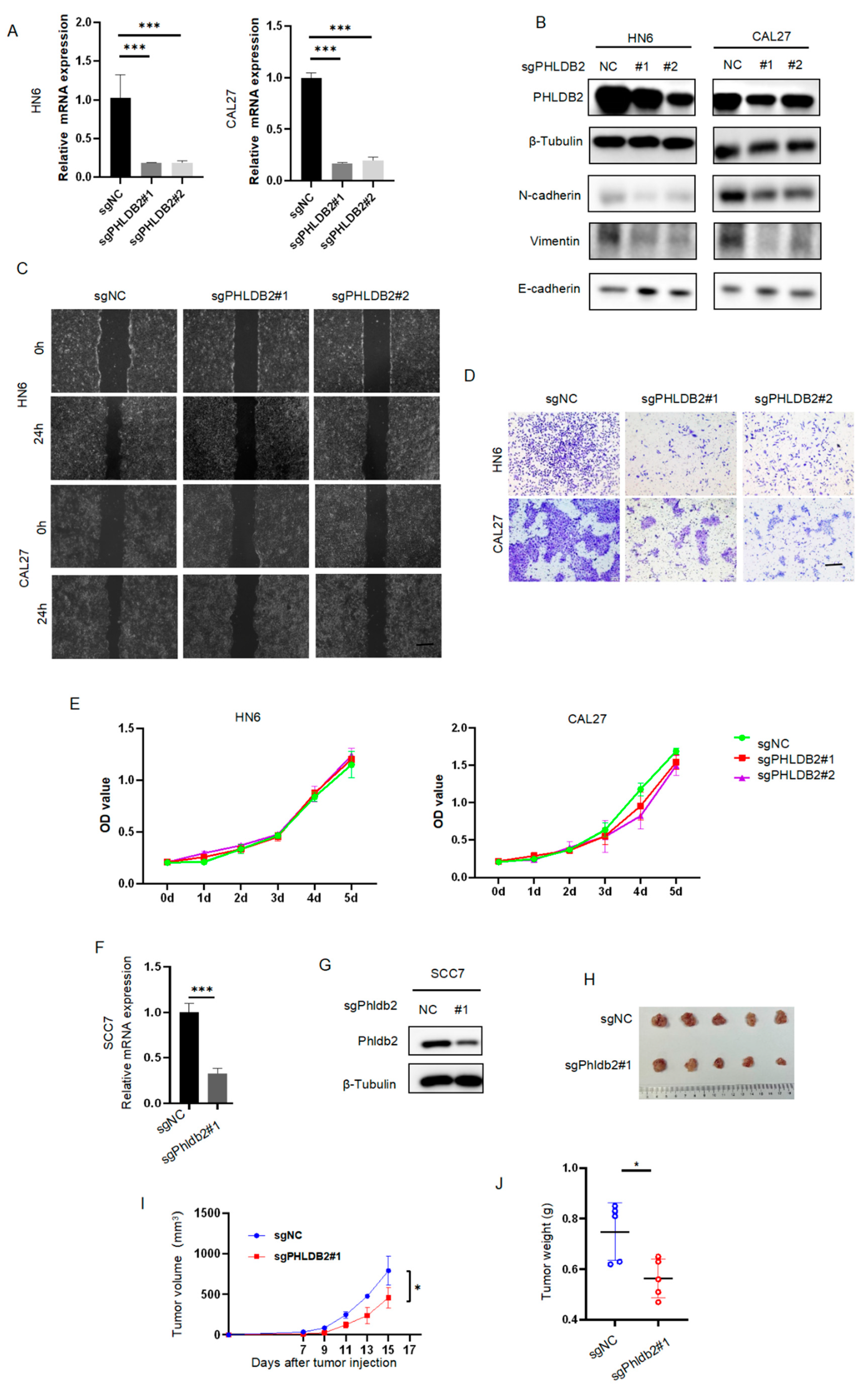
3.2. PHLDB2 Promotes EMT and Tumorigenicity of HNSCC
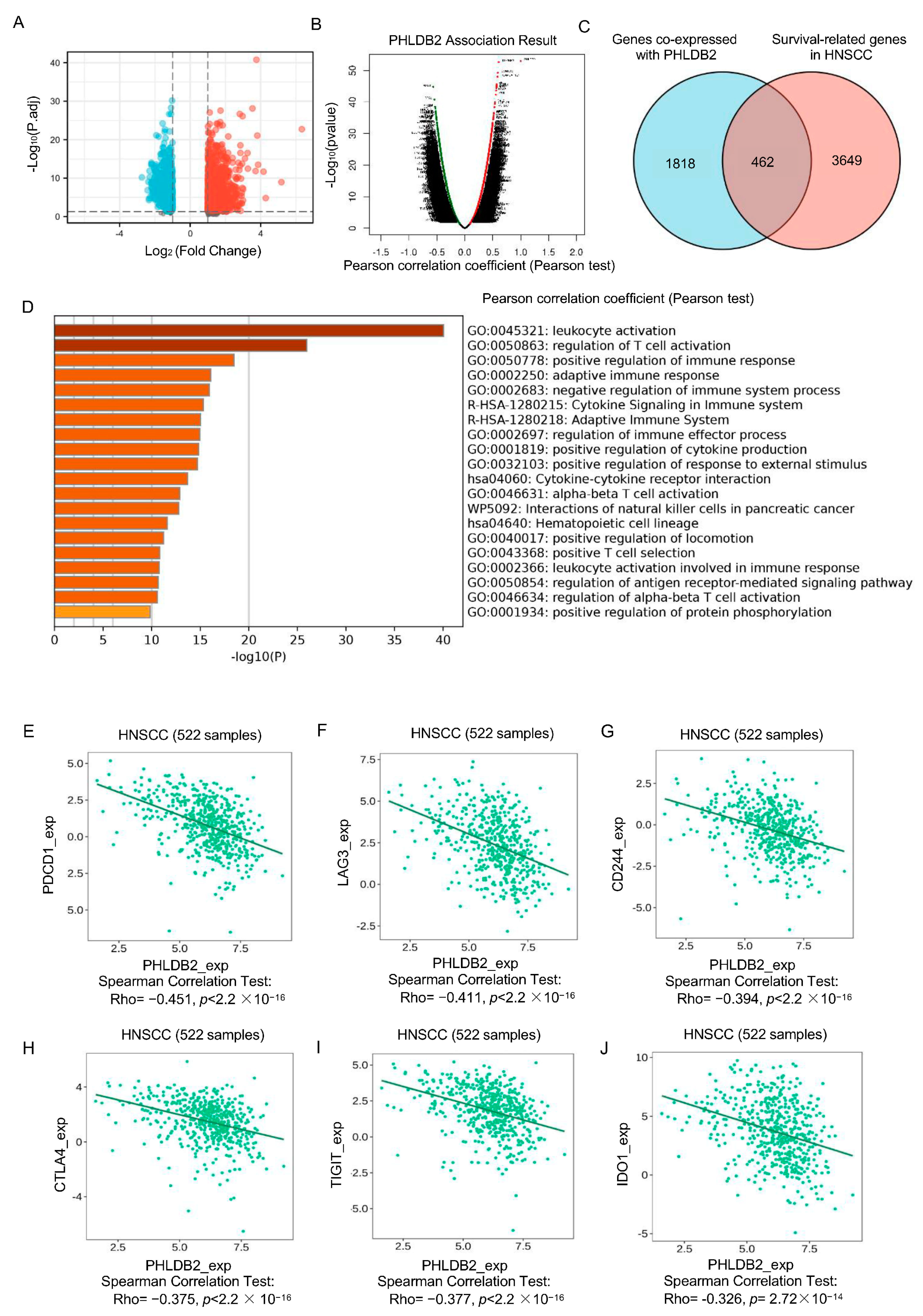
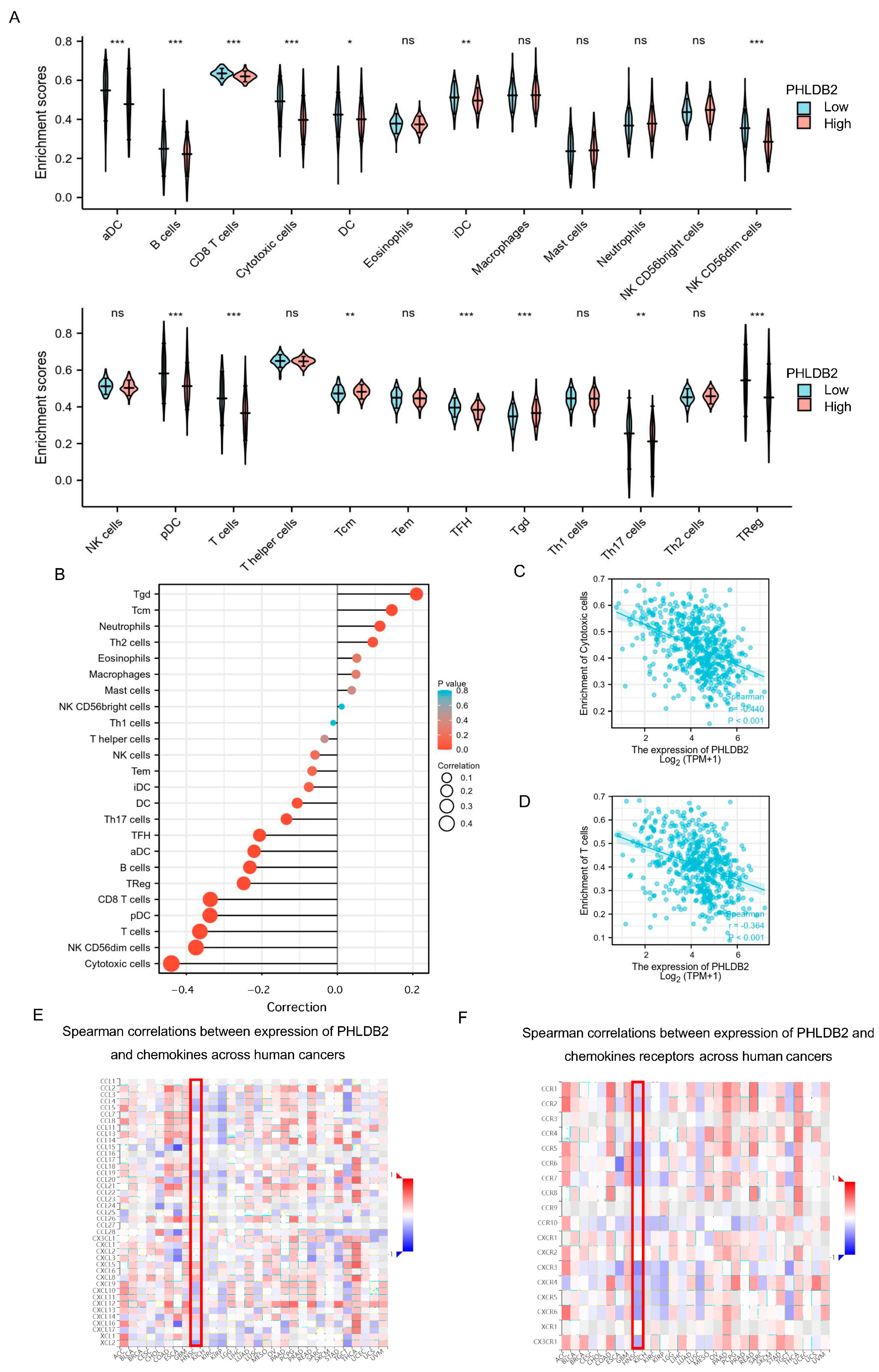
3.3. Elevated PHLDB2 Is Closely Related to Immune Regulation in HNSCC
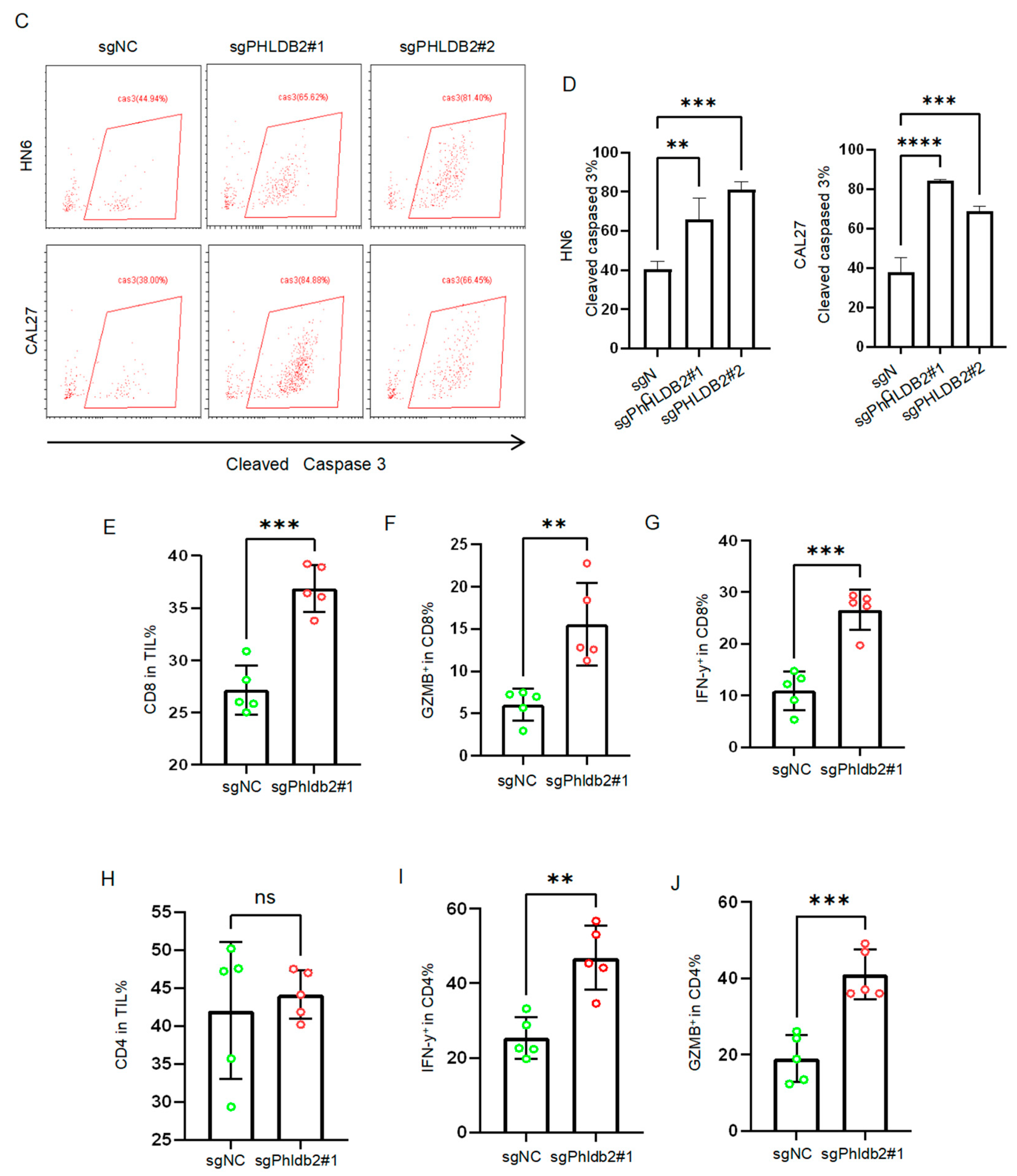
3.4. Targeting PHLDB2 Exerts an Anti-Tumor Effect in HNSCC by Enhancing the Anti-Tumor Function of T Cells
4. Discussions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Cramer, J.D.; Burtness, B.; Le, Q.T.; Ferris, R.L. The changing therapeutic landscape of head and neck cancer. Nat. Rev. Clin. Oncol. 2019, 16, 669–683. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Hedberg, M.L.; Goh, G.; Chiosea, S.I.; Bauman, J.E.; Freilino, M.L.; Zeng, Y.; Wang, L.; Diergaarde, B.B.; Gooding, W.E.; Lui, V.W.; et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J. Clin. Investig. 2016, 126, 1606. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Economopoulou, P.; Agelaki, S.; Perisanidis, C.; Giotakis, E.I.; Psyrri, A. The promise of immunotherapy in head and neck squamous cell carcinoma. Ann. Oncol. 2016, 27, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Ling, D.C.; Bakkenist, C.J.; Ferris, R.L.; Clump, D.A. Role of Immunotherapy in Head and Neck Cancer. Semin. Radiat. Oncol. 2018, 28, 12–16. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Page, D.B.; Postow, M.A.; Callahan, M.K.; Allison, J.P.; Wolchok, J.D. Immune modulation in cancer with antibodies. Annu. Rev. Med. 2014, 65, 185–202. [Google Scholar] [CrossRef]
- Hanna, B.S.; Llaó-Cid, L.; Iskar, M.; Roessner, P.M.; Klett, L.C.; Wong, J.K.L.; Paul, Y.; Ioannou, N.; Öztürk, S.; Mack, N.; et al. Interleukin-10 receptor signaling promotes the maintenance of a PD-1(int) TCF-1(+) CD8(+) T cell population that sustains anti-tumor immunity. Immunity 2021, 54, 2825–2841.e2810. [Google Scholar] [CrossRef]
- Virassamy, B.; Caramia, F.; Savas, P.; Sant, S.; Wang, J.; Christo, S.N.; Byrne, A.; Clarke, K.; Brown, E.; Teo, Z.L.; et al. Intratumoral CD8(+) T cells with a tissue-resident memory phenotype mediate local immunity and immune checkpoint responses in breast cancer. Cancer Cell. 2023, 41, 585–601.e588. [Google Scholar] [CrossRef]
- Lim, B.C.; Matsumoto, S.; Yamamoto, H.; Mizuno, H.; Kikuta, J.; Ishii, M.; Kikuchi, A. Prickle1 promotes focal adhesion disassembly in cooperation with the CLASP-LL5β complex in migrating cells. J. Cell Sci. 2016, 129, 3115–3129. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, T.; Ma, T.; Cao, T.; Yu, Z. Oncogenic effect of PHLDB2 is associated with epithelial-mesenchymal transition and E-cadherin regulation in colorectal cancer. Cancer Cell Int. 2019, 19, 184. [Google Scholar] [CrossRef]
- Hotta, A.; Kawakatsu, T.; Nakatani, T.; Sato, T.; Matsui, C.; Sukezane, T.; Akagi, T.; Hamaji, T.; Grigoriev, I.; Akhmanova, A.; et al. Laminin-based cell adhesion anchors microtubule plus ends to the epithelial cell basal cortex through LL5alpha/beta. J. Cell Biol. 2010, 189, 901–917. [Google Scholar] [CrossRef]
- Luo, M.; Huang, Z.; Yang, X.; Chen, Y.; Jiang, J.; Zhang, L.; Zhou, L.; Qin, S.; Jin, P.; Fu, S.; et al. PHLDB2 Mediates Cetuximab Resistance via Interacting With EGFR in Latent Metastasis of Colorectal Cancer. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1223–1242. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Zhang, J. NOTCH3, a crucial target of miR-491-5p/miR-875-5p, promotes gastric carcinogenesis by upregulating PHLDB2 expression and activating Akt pathway. Oncogene 2021, 40, 1578–1594. [Google Scholar] [CrossRef] [PubMed]
- Carles, A.; Millon, R.; Cromer, A.; Ganguli, G.; Lemaire, F.; Young, J.; Wasylyk, C.; Muller, D.; Schultz, I.; Rabouel, Y.; et al. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene 2006, 25, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Feng, M.; Bai, L.; Liao, W.; Zhou, K.; Zhang, M.; Wu, Q.; Wen, F.; Lei, W.; Zhang, P.; et al. Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer. J. Transl. Med. 2021, 19, 391. [Google Scholar] [CrossRef] [PubMed]
- Astro, V.; Chiaretti, S.; Magistrati, E.; Fivaz, M.; de Curtis, I. Liprin-α1, ERC1 and LL5 define polarized and dynamic structures that are implicated in cell migration. J. Cell Sci. 2014, 127, 3862–3876. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, S.; Liu, Y.; Li, C.; Guo, T.; Zhang, S.; Li, Z.; Jiao, Z.; Sun, H.; Zhang, Y.; et al. NR2F1-AS1/miR-190a/PHLDB2 Induces the Epithelial-Mesenchymal Transformation Process in Gastric Cancer by Promoting Phosphorylation of AKT3. Front. Cell Dev. Biol. 2021, 9, 688949. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Schuhwerk, H. Dynamic EMT: A multi-tool for tumor progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef] [PubMed]
- Taki, M.; Abiko, K.; Ukita, M.; Murakami, R.; Yamanoi, K.; Yamaguchi, K. Tumor Immune Microenvironment during Epithelial-Mesenchymal Transition. Clin. Cancer Res. 2021, 27, 4669–4679. [Google Scholar] [CrossRef] [PubMed]
- Chockley, P.J.; Chen, J.; Chen, G.; Beer, D.G.; Standiford, T.J.; Keshamouni, V.G. Epithelial-mesenchymal transition leads to NK cell-mediated metastasis-specific immunosurveillance in lung cancer. J. Clin. Investig. 2018, 128, 1384–1396. [Google Scholar] [CrossRef]
- Imai, D.; Yoshizumi, T.; Okano, S.; Itoh, S.; Ikegami, T.; Harada, N.; Aishima, S.; Oda, Y.; Maehara, Y. IFN-γ Promotes Epithelial-Mesenchymal Transition and the Expression of PD-L1 in Pancreatic Cancer. J. Surg. Res. 2019, 240, 115–123. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhan, H. Communication between EMT and PD-L1 signaling: New insights into tumor immune evasion. Cancer Lett. 2020, 468, 72–81. [Google Scholar] [CrossRef]
- Lou, Y.; Diao, L.; Cuentas, E.R.; Denning, W.L.; Chen, L.; Fan, Y.H.; Byers, L.A.; Wang, J.; Papadimitrakopoulou, V.A.; Behrens, C.; et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3630–3642. [Google Scholar] [CrossRef]
- Kim, S.; Koh, J.; Kim, M.Y.; Kwon, D.; Go, H.; Kim, Y.A.; Jeon, Y.K.; Chung, D.H. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Human Pathol. 2016, 58, 7–14. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Zheng, Q.; Lu, Z.; Chen, Y.; Shen, D.; Xue, D.; Jiang, M.; Ding, L.; Zhang, J.; et al. Oncometabolite L-2-hydroxyglurate directly induces vasculogenic mimicry through PHLDB2 in renal cell carcinoma. Int. J. Cancer 2021, 148, 1743–1755. [Google Scholar] [CrossRef]
- Chen, G.; Zhou, T.; Li, Y.; Yu, Z.; Sun, L. p53 target miR-29c-3p suppresses colon cancer cell invasion and migration through inhibition of PHLDB2. Biochem. Biophys. Res. Commun. 2017, 487, 90–95. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef]
- Prokhnevska, N.; Cardenas, M.A.; Valanparambil, R.M.; Sobierajska, E.; Barwick, B.G.; Jansen, C.; Reyes Moon, A.; Gregorova, P.; delBalzo, L.; Greenwald, R.; et al. CD8(+) T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity 2023, 56, 107–124.e105. [Google Scholar] [CrossRef]
- Azimi, F.; Scolyer, R.A.; Rumcheva, P.; Moncrieff, M.; Murali, R.; McCarthy, S.W.; Saw, R.P.; Thompson, J.F. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol. 2012, 30, 2678–2683. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ruan, Y.Q.; Qu, L.H.; Li, Z.H.; Xie, C.; Pan, Y.Q.; Li, H.F.; Li, D.B. Prognostic Modeling of Lung Adenocarcinoma Based on Hypoxia and Ferroptosis-Related Genes. J. Oncol. 2022, 2022, 1022580. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, I.; Akutsu, Y.; Murakami, K.; Akanuma, N.; Isozaki, Y.; Maruyama, T.; Toyozumi, T.; Matsumoto, Y.; Suito, H.; Takahashi, M.; et al. Histone Demethylase LSD1 Inhibitors Prevent Cell Growth by Regulating Gene Expression in Esophageal Squamous Cell Carcinoma Cells. Ann. Surg. Oncol. 2016, 23, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.I.; Faivre, S.; Licitra, L.; Machiels, J.P.; Vermorken, J.B.; Bruzzi, P.; Gruenwald, V.; Giglio, R.E.; Leemans, C.R.; Seiwert, T.Y.; et al. Comparative analysis of the phase III clinical trials of anti-PD1 monotherapy in head and neck squamous cell carcinoma patients (CheckMate 141 and KEYNOTE 040). J. Immunother. Cancer 2019, 7, 96. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.T.; Xia, R.H.; Zhu, D.W.; Dou, S.J.; Zhu, G.P. A pilot study of neoadjuvant combination of anti-PD-1 camrelizumab and VEGFR2 inhibitor apatinib for locally advanced resectable oral squamous cell carcinoma. Nat. Commun. 2022, 13, 5378. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Mudianto, T.; Ma, X.; Riley, R.; Uppaluri, R. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity, and Circumvents Anti-PD-1 Resistance in Head and Neck Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 290–300. [Google Scholar] [CrossRef]
- Dorta-Estremera, S.; Hegde, V.L.; Slay, R.B.; Sun, R.; Yanamandra, A.V.; Nicholas, C.; Nookala, S.; Sierra, G.; Curran, M.A.; Sastry, K.J. Targeting interferon signaling and CTLA-4 enhance the therapeutic efficacy of anti-PD-1 immunotherapy in preclinical model of HPV(+) oral cancer. J. Immunother. Cancer. 2019, 7, 252. [Google Scholar] [CrossRef]
- Upadhaya, S.; Neftelino, S.T.; Hodge, J.P.; Oliva, C.; Campbell, J.R.; Yu, J.X. Combinations take centre stage in PD1/PDL1 inhibitor clinical trials. Nat. Rev. Drug Discov. 2021, 20, 168–169. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Larkin, J.; Tabernero, J.; Bonini, C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet 2021, 397, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell. 2020, 37, 443–455. [Google Scholar] [CrossRef] [PubMed]
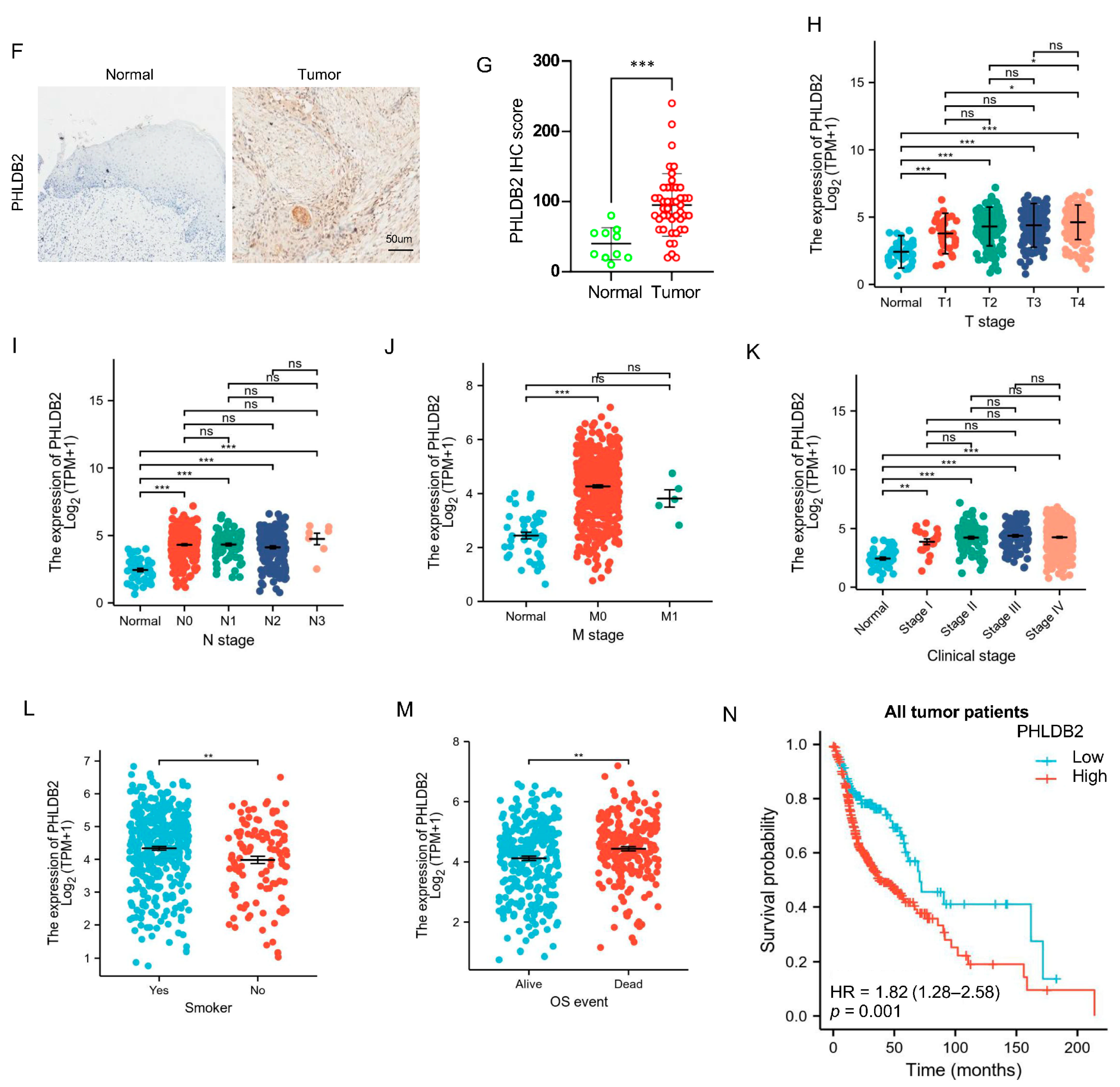

| Characteristic | Low Expression of PHLDB2 | High Expression of PHLDB2 | p-Value |
|---|---|---|---|
| n | 251 | 251 | |
| T stage, n (%) | 0.024 | ||
| T1 | 21 (4.3%) | 12 (2.5%) | |
| T2 | 81 (16.6%) | 63 (12.9%) | |
| T3 | 66 (13.6%) | 65 (13.3%) | |
| T4 | 75 (15.4%) | 104 (21.4%) | |
| N stage, n (%) | 0.780 | ||
| N0 | 120 (25%) | 119 (24.8%) | |
| N1 | 40 (8.3%) | 40 (8.3%) | |
| N2 | 77 (16%) | 77 (16%) | |
| N3 | 2 (0.4%) | 5 (1%) | |
| M stage, n (%) | 0.216 | ||
| M0 | 234 (49.1%) | 238 (49.9%) | |
| M1 | 4 (0.8%) | 1 (0.2%) | |
| Clinical stage, n (%) | 0.524 | ||
| Stage I | 12 (2.5%) | 7 (1.4%) | |
| Stage II | 51 (10.5%) | 44 (9%) | |
| Stage III | 50 (10.2%) | 52 (10.7%) | |
| Stage IV | 131 (26.8%) | 141 (28.9%) | |
| Gender, n (%) | 0.034 | ||
| Female | 78 (15.5%) | 56 (11.2%) | |
| Male | 173 (34.5%) | 195 (38.8%) | |
| Age, n (%) | 0.395 | ||
| ≤60 | 117 (23.4%) | 128 (25.5%) | |
| >60 | 133 (26.5%) | 123 (24.6%) | |
| Histologic grade, n (%) | <0.001 | ||
| G1 | 43 (8.9%) | 19 (3.9%) | |
| G2 | 125 (25.9%) | 175 (36.2%) | |
| G3 | 70 (14.5%) | 49 (10.1%) | |
| G4 | 1 (0.2%) | 1 (0.2%) | |
| Smoker, n (%) | 0.015 | ||
| No | 67 (13.6%) | 44 (8.9%) | |
| Yes | 178 (36.2%) | 203 (41.3%) | |
| Alcohol history, n (%) | 0.106 | ||
| No | 89 (18.1%) | 69 (14.1%) | |
| Yes | 160 (32.6%) | 173 (35.2%) | |
| Age, median (IQR) | 61 (55, 69) | 60 (53, 67) | 0.187 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Wang, Z.; Liang, H.; Liu, X.; Liu, H.; Zhuang, Z.; Hou, J. Depletion of PHLDB2 Suppresses Epithelial–Mesenchymal Transition and Enhances Anti-Tumor Immunity in Head and Neck Squamous Cell Carcinoma. Biomolecules 2024, 14, 232. https://doi.org/10.3390/biom14020232
Li H, Wang Z, Liang H, Liu X, Liu H, Zhuang Z, Hou J. Depletion of PHLDB2 Suppresses Epithelial–Mesenchymal Transition and Enhances Anti-Tumor Immunity in Head and Neck Squamous Cell Carcinoma. Biomolecules. 2024; 14(2):232. https://doi.org/10.3390/biom14020232
Chicago/Turabian StyleLi, Hongyu, Ziyi Wang, Huiting Liang, Xiaoyong Liu, Haichao Liu, Zehang Zhuang, and Jinsong Hou. 2024. "Depletion of PHLDB2 Suppresses Epithelial–Mesenchymal Transition and Enhances Anti-Tumor Immunity in Head and Neck Squamous Cell Carcinoma" Biomolecules 14, no. 2: 232. https://doi.org/10.3390/biom14020232





