Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of Cells for Immunofluorescence Microscopy
2.3. Antibodies for Immunofluorescence Microscopy and Western Blot Analysis
2.4. Western Blot Analysis
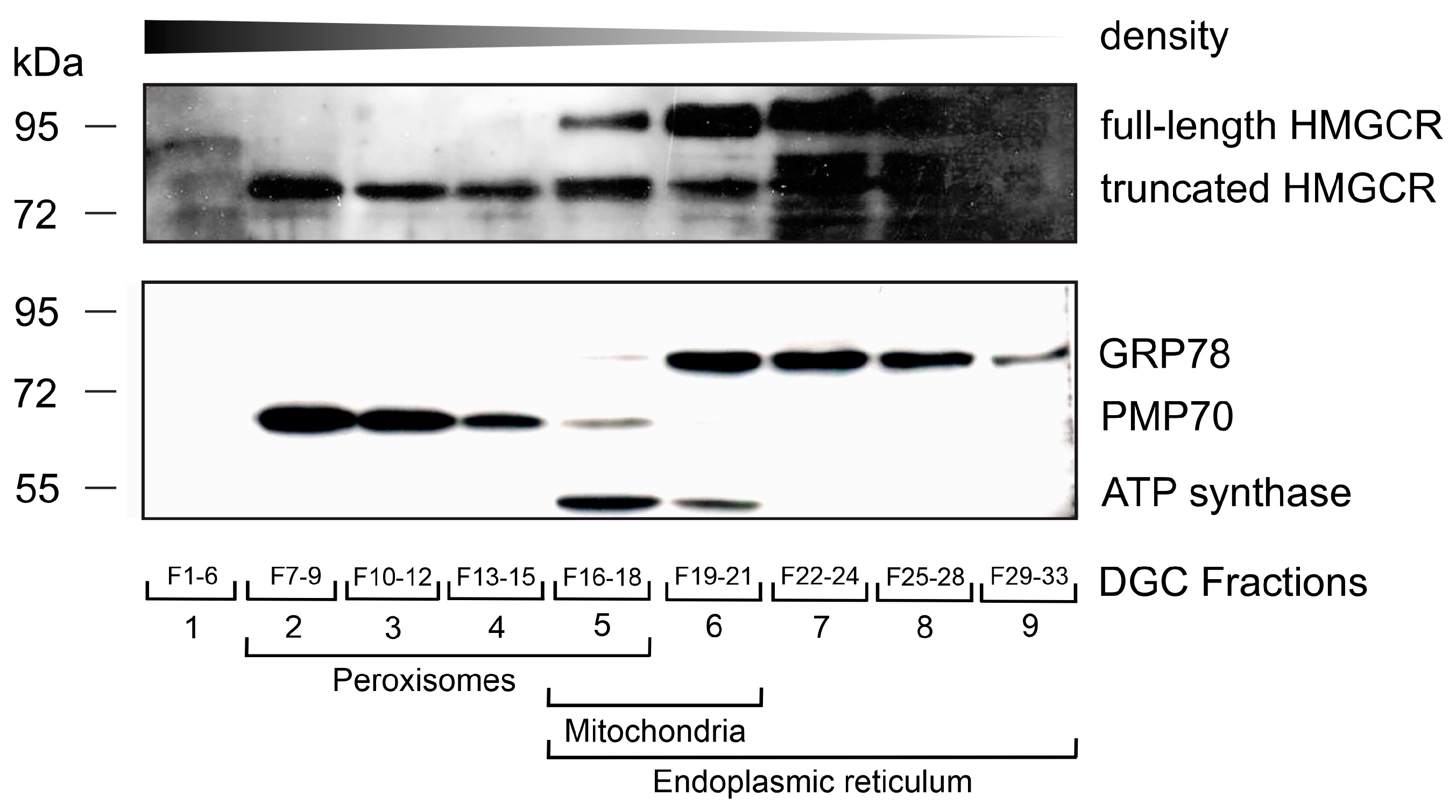
2.5. Density Gradient Centrifugation for Organelle Fractionation
2.6. PTS2 Reporter Constructs and Analysis of Peroxisomal Targeting in COS-7 Cells
2.7. Statistical Analysis
3. Results
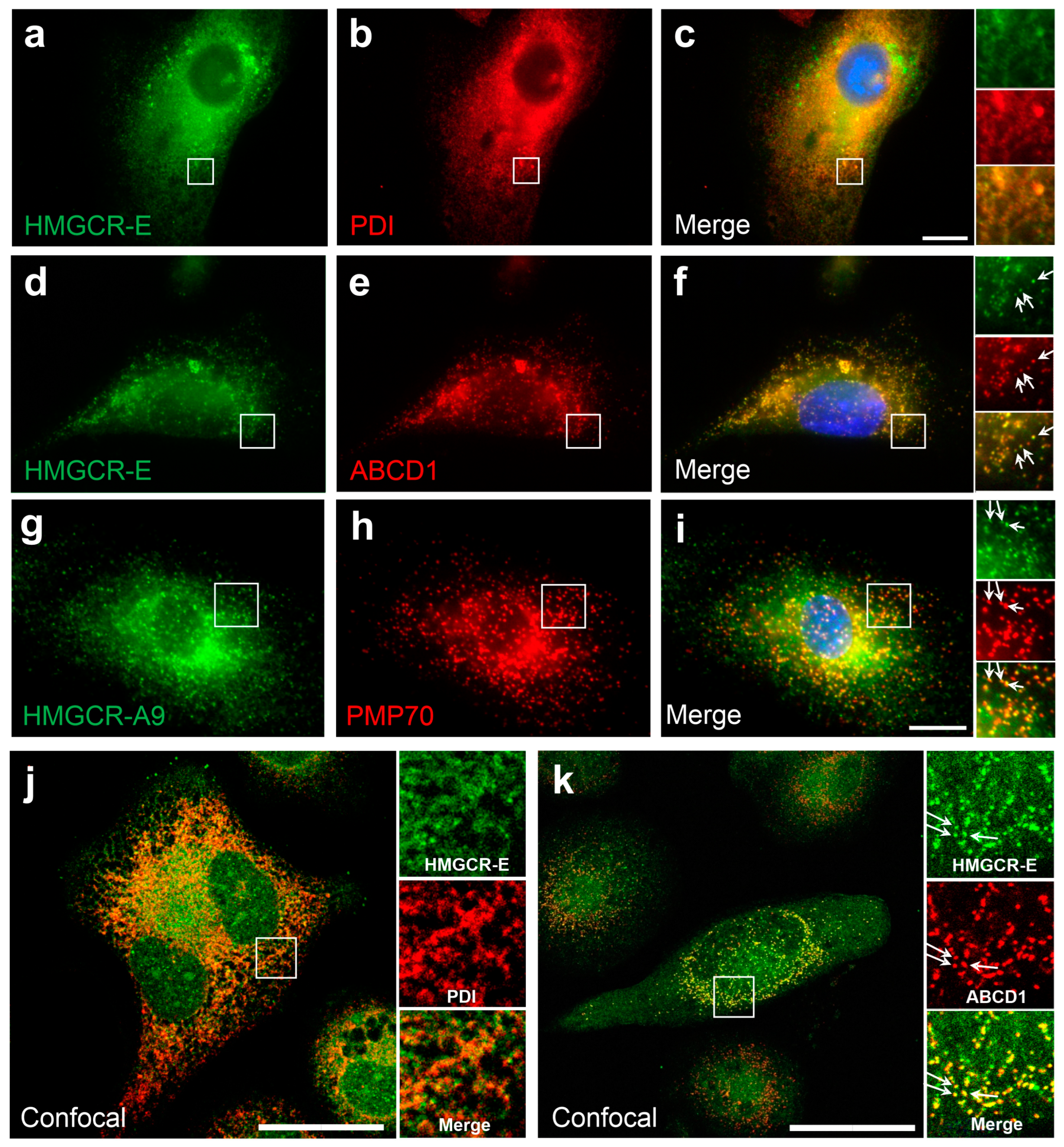
3.1. HMG-CoA Reductase Is Bilocalized to ER and Peroxisomes in Human Differentiated THP-1 Cells under Conditions of Low Cholesterol and Lovastatin Treatment
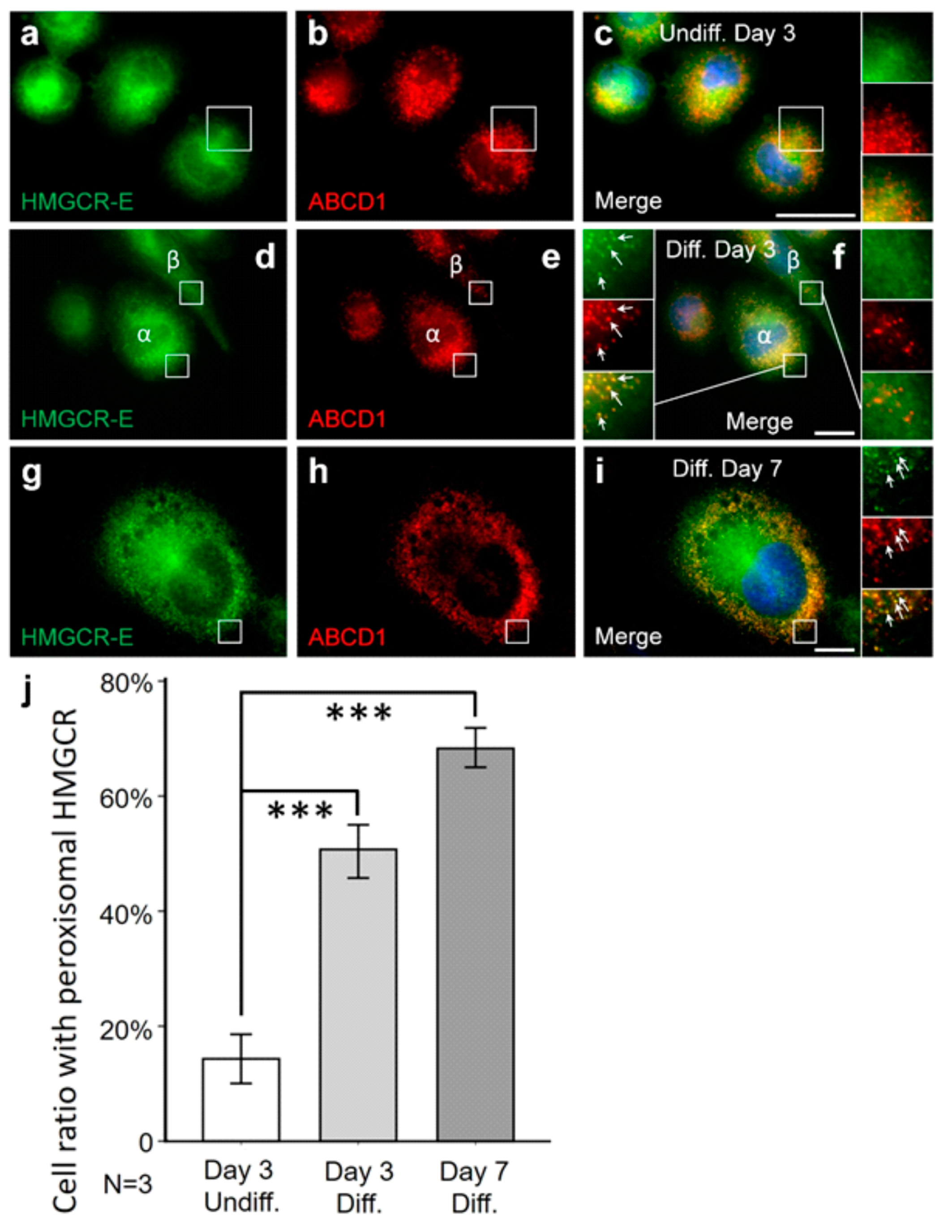
3.2. The Fraction of Peroxisomal HMGCR Increases with the Differentiation State of THP-1 Cells
3.3. A Truncated HMGCR Is Located to Peroxisomes
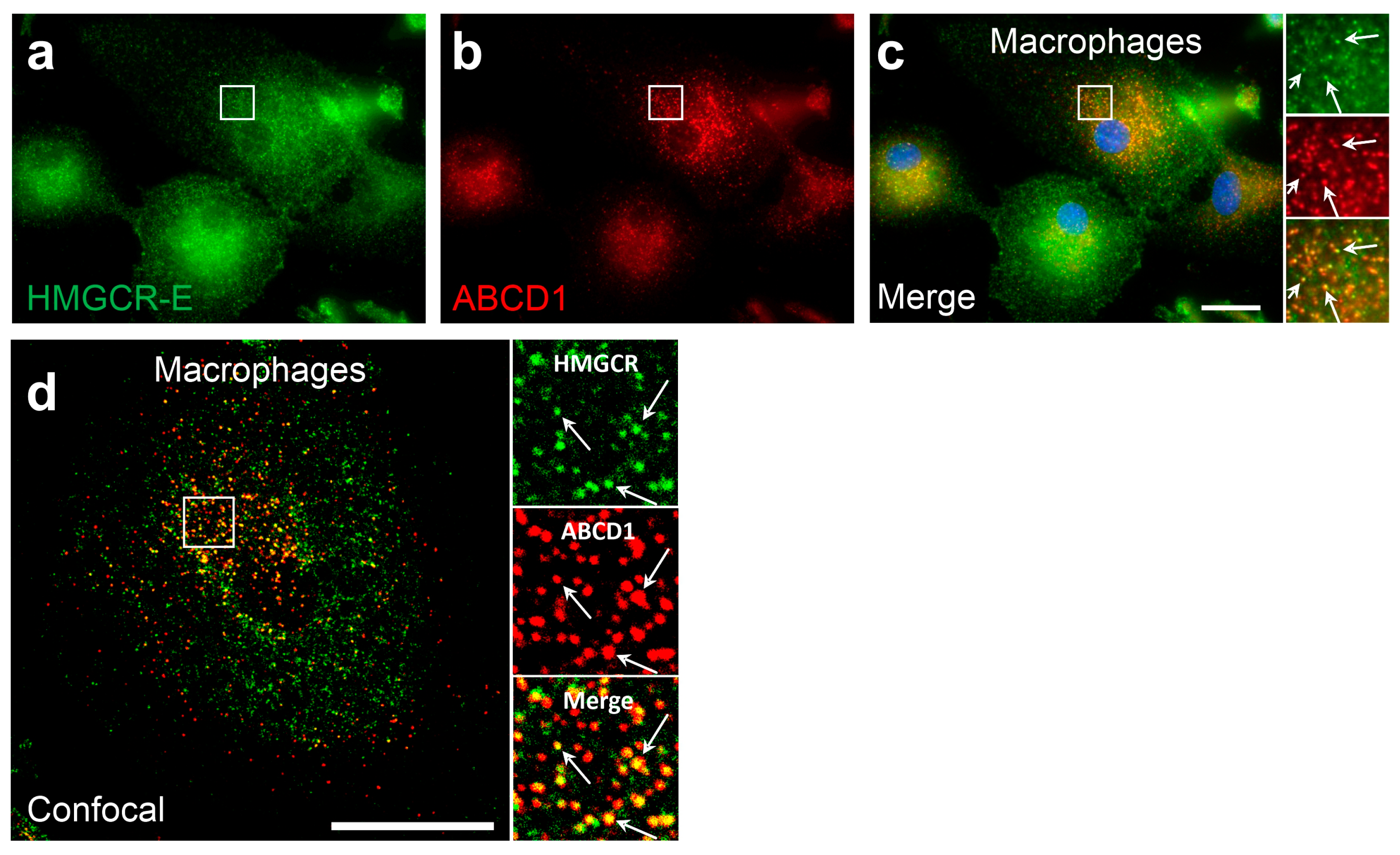
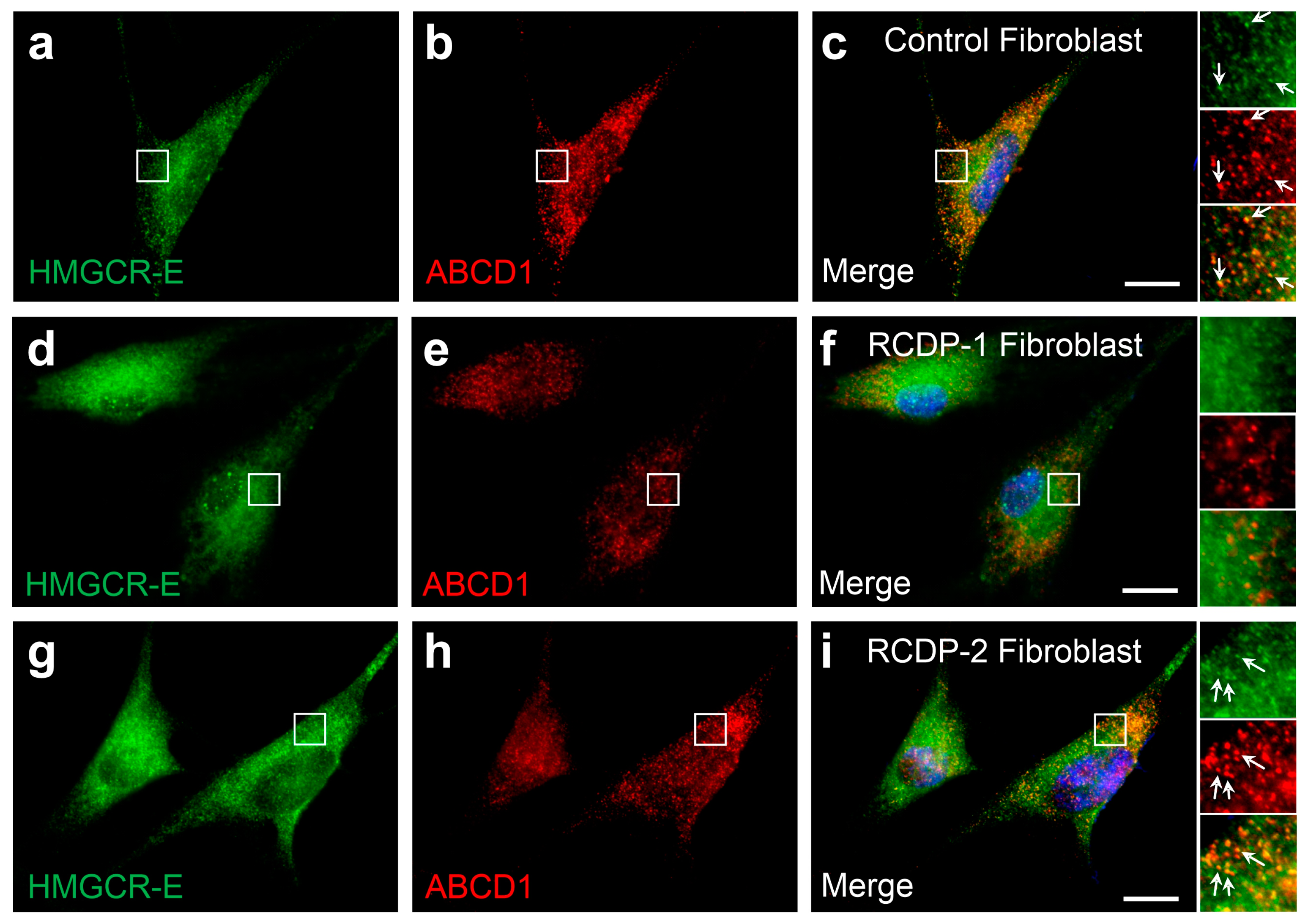
3.4. The Percentage of Cells with Detectable Peroxisomal HMGCR under Low-Cholesterol Conditions and Lovastatin Treatment Is Cell-Type-Specific
3.5. PEX7 Is Required for the Peroxisomal Localization of Human HMGCR
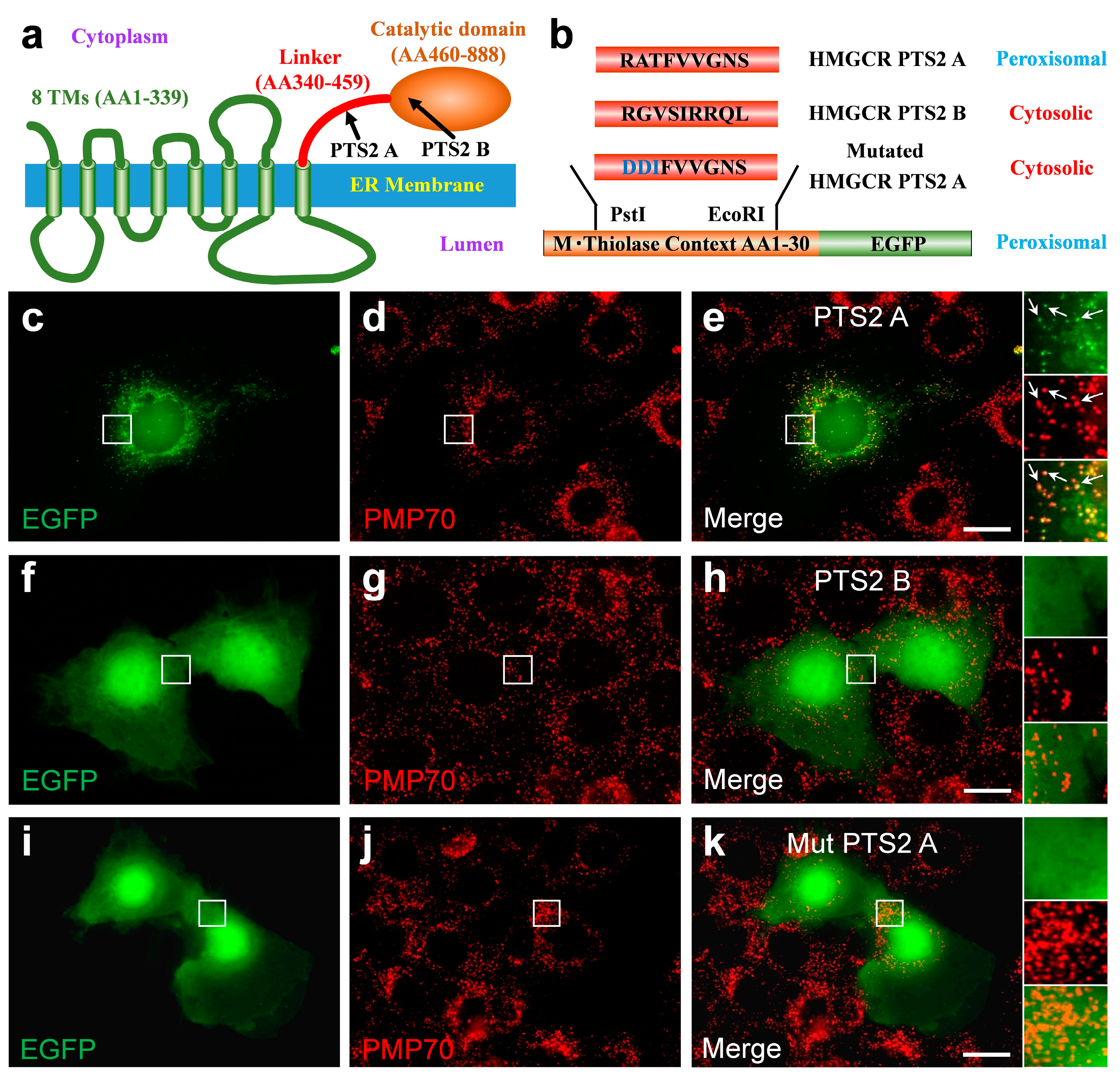
3.6. The C-Terminal Domain of HMGCR Contains a Functional PTS2 Close to Its N-Terminus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Hinson, D.D.; Chambliss, K.L.; Toth, M.J.; Tanaka, R.D.; Gibson, K.M. Post-translational regulation of mevalonate kinase by intermediates of the cholesterol and nonsterol isoprene biosynthetic pathways. J. Lipid Res. 1997, 38, 2216–2223. [Google Scholar] [CrossRef]
- Edwards, P.A.; Ericsson, J. Sterols and isoprenoids: Signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 1999, 68, 157–185. [Google Scholar] [CrossRef]
- Morales-Rosado, J.A.; Schwab, T.L.; Macklin-Mantia, S.K.; Foley, A.R.; Pinto, E.V.F.; Pehlivan, D.; Donkervoort, S.; Rosenfeld, J.A.; Boyum, G.E.; Hu, Y.; et al. Bi-allelic variants in hmgcr cause an autosomal-recessive progressive limb-girdle muscular dystrophy. Am. J. Hum. Genet. 2023, 110, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Meigs, T.E.; Roseman, D.S.; Simoni, R.D. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme a reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J. Biol. Chem. 1996, 271, 7916–7922. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.J.; Sargeant, T.E.; Watson, J.A. Inhibition of 3-hydroxy-3-methylglutaryl coenzyme a reductase activity in hepatoma tissue culture cells by pure cholesterol and several cholesterol derivatives. Evidence supporting two distinct mechanisms.20l. J. Biol. Chem. 1976, 251, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.A.; Lan, S.F.; Tanaka, R.D.; Fogelman, A.M. Mevalonolactone inhibits the rate of synthesis and enhances the rate of degradation of 3-hydroxy-3-methylglutaryl coenzyme a reductase in rat hepatocytes. J. Biol. Chem. 1983, 258, 7272–7275. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. Cholesterol feedback: From schoenheimer’s bottle to scap’s meladl. J. Lipid Res. 2009, 50, S15–S27. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. The ldl receptor. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of hmg-coa reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Tobert, J.A. Lovastatin and beyond: The history of the hmg-coa reductase inhibitors. Nat. Rev. Drug Discov. 2003, 2, 517–526. [Google Scholar] [CrossRef]
- Pappu, A.S.; Illingworth, D.R. Contrasting effects of lovastatin and cholestyramine on low-density lipoprotein cholesterol and 24-hour urinary mevalonate excretion in patients with heterozygous familial hypercholesterolemia. J. Lab. Clin. Med. 1989, 114, 554–562. [Google Scholar]
- Brown, M.S.; Goldstein, J.L. Multivalent feedback regulation of hmg coa reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 1980, 21, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Simoni, R.D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme a reductase, an integral glycoprotein of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1984, 81, 1674–1678. [Google Scholar] [CrossRef] [PubMed]
- Chin, D.J.; Gil, G.; Russell, D.W.; Liscum, L.; Luskey, K.L.; Basu, S.K.; Okayama, H.; Berg, P.; Goldstein, J.L.; Brown, M.S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme a reductase, a glycoprotein of endoplasmic reticulum. Nature 1984, 308, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Liscum, L.; Finer-Moore, J.; Stroud, R.M.; Luskey, K.L.; Brown, M.S.; Goldstein, J.L. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme a reductase, a glycoprotein of the endoplasmic reticulum. J. Biol. Chem. 1985, 260, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Luskey, K.L.; Stevens, B. Human 3-hydroxy-3-methylglutaryl coenzyme a reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J. Biol. Chem. 1985, 260, 10271–10277. [Google Scholar] [CrossRef] [PubMed]
- Roitelman, J.; Olender, E.H.; Bar-Nun, S.; Dunn, W.A., Jr.; Simoni, R.D. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme a reductase: Implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol. 1992, 117, 959–973. [Google Scholar] [CrossRef]
- Friesen, J.A.; Rodwell, V.W. The 3-hydroxy-3-methylglutaryl coenzyme-a (hmg-coa) reductases. Genome Biol. 2004, 5, 248. [Google Scholar] [CrossRef] [PubMed]
- Werner, E.D.; Brodsky, J.L.; McCracken, A.A. Proteasome-dependent endoplasmic reticulum-associated protein degradation: An unconventional route to a familiar fate. Proc. Natl. Acad. Sci. USA 1996, 93, 13797–13801. [Google Scholar] [CrossRef]
- Jo, Y.; Hartman, I.Z.; DeBose-Boyd, R.A. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl coa reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 2013, 24, 169–183. [Google Scholar] [CrossRef]
- Gil, G.; Faust, J.R.; Chin, D.J.; Goldstein, J.L.; Brown, M.S. Membrane-bound domain of hmg coa reductase is required for sterol-enhanced degradation of the enzyme. Cell 1985, 41, 249–258. [Google Scholar] [CrossRef]
- Roitelman, J.; Simoni, R.D. Distinct sterol and nonsterol signals for the regulated degradation of 3-hydroxy-3-methylglutaryl-coa reductase. J. Biol. Chem. 1992, 267, 25264–25273. [Google Scholar] [CrossRef]
- Inoue, S.; Bar-Nun, S.; Roitelman, J.; Simoni, R.D. Inhibition of degradation of 3-hydroxy-3-methylglutaryl-coenzyme a reductase in vivo by cysteine protease inhibitors. J. Biol. Chem. 1991, 266, 13311–13317. [Google Scholar] [CrossRef]
- McGee, T.P.; Cheng, H.H.; Kumagai, H.; Omura, S.; Simoni, R.D. Degradation of 3-hydroxy-3-methylglutaryl-coa reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J. Biol. Chem. 1996, 271, 25630–25638. [Google Scholar] [CrossRef]
- Ravid, T.; Doolman, R.; Avner, R.; Harats, D.; Roitelman, J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme a reductase. J. Biol. Chem. 2000, 275, 35840–35847. [Google Scholar] [CrossRef]
- Sever, N.; Yang, T.; Brown, M.S.; Goldstein, J.L.; DeBose-Boyd, R.A. Accelerated degradation of hmg coa reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 2003, 11, 25–33. [Google Scholar] [CrossRef]
- Johnson, B.M.; DeBose-Boyd, R.A. Underlying mechanisms for sterol-induced ubiquitination and er-associated degradation of hmg coa reductase. Semin. Cell Dev. Biol. 2018, 81, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Hartman, I.Z.; Liu, P.; Zehmer, J.K.; Luby-Phelps, K.; Jo, Y.; Anderson, R.G.; DeBose-Boyd, R.A. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme a reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. J. Biol. Chem. 2010, 285, 19288–19298. [Google Scholar] [CrossRef] [PubMed]
- Keller, G.A.; Barton, M.C.; Shapiro, D.J.; Singer, S.J. 3-hydroxy-3-methylglutaryl-coenzyme a reductase is present in peroxisomes in normal rat liver cells. Proc. Natl. Acad. Sci. USA 1985, 82, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Li, A.C.; Tanaka, R.D.; Callaway, K.; Fogelman, A.M.; Edwards, P.A. Localization of 3-hydroxy-3-methylglutaryl coa reductase and 3-hydroxy-3-methylglutaryl coa synthase in the rat liver and intestine is affected by cholestyramine and mevinolin. J. Lipid Res. 1988, 29, 781–796. [Google Scholar] [CrossRef]
- Kovacs, W.J.; Faust, P.L.; Keller, G.A.; Krisans, S.K. Purification of brain peroxisomes and localization of 3-hydroxy-3-methylglutaryl coenzyme a reductase. Eur. J. Biochem. 2001, 268, 4850–4859. [Google Scholar] [CrossRef]
- Wanders, R.J.; Ferdinandusse, S.; Brites, P.; Kemp, S. Peroxisomes, lipid metabolism and lipotoxicity. Biochim. Biophys. Acta 2010, 1801, 272–280. [Google Scholar] [CrossRef]
- Islinger, M.; Grille, S.; Fahimi, H.D.; Schrader, M. The peroxisome: An update on mysteries. Histochem. Cell Biol. 2012, 137, 547–574. [Google Scholar] [CrossRef]
- Wanders, R.J.; Waterham, H.R. Peroxisomal disorders i: Biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 2005, 67, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.; Waterham, H.R. Peroxisomal disorders: The single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta 2006, 1763, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Argyriou, C.; D’Agostino, M.D.; Braverman, N. Peroxisome biogenesis disorders. Transl. Sci. Rare Dis. 2016, 1, 111–144. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta 2016, 1863, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A.; Baes, M.; Ribeiro, D.; Ferdinandusse, S.; Waterham, H.R. The physiological functions of human peroxisomes. Physiol. Rev. 2023, 103, 957–1024. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.; Waterham, H.R. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 2006, 75, 295–332. [Google Scholar] [CrossRef]
- Berger, J.; Dorninger, F.; Forss-Petter, S.; Kunze, M. Peroxisomes in brain development and function. Biochim. Biophys. Acta 2016, 1863, 934–955. [Google Scholar] [CrossRef] [PubMed]
- Dorninger, F.; Werner, E.R.; Berger, J.; Watschinger, K. Regulation of plasmalogen metabolism and traffic in mammals: The fog begins to lift. Front. Cell Dev. Biol. 2022, 10, 946393. [Google Scholar] [CrossRef] [PubMed]
- Dorninger, F.; Forss-Petter, S.; Wimmer, I.; Berger, J. Plasmalogens, platelet-activating factor and beyond—Ether lipids in signaling and neurodegeneration. Neurobiol. Dis. 2020, 145, 105061. [Google Scholar] [CrossRef] [PubMed]
- Weinhofer, I.; Buda, A.; Kunze, M.; Palfi, Z.; Traunfellner, M.; Hesse, S.; Villoria-Gonzalez, A.; Hofmann, J.; Hametner, S.; Regelsberger, G.; et al. Peroxisomal very long-chain fatty acid transport is targeted by herpesviruses and the antiviral host response. Commun. Biol. 2022, 5, 944. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.R.; Marques, M.; Ramos, B.; Kagan, J.C.; Ribeiro, D. Emerging roles of peroxisomes in viral infections. Trends Cell Biol. 2022, 32, 124–139. [Google Scholar] [CrossRef]
- Keller, G.A.; Pazirandeh, M.; Krisans, S. 3-hydroxy-3-methylglutaryl coenzyme a reductase localization in rat liver peroxisomes and microsomes of control and cholestyramine-treated animals: Quantitative biochemical and immunoelectron microscopical analyses. J. Cell Biol. 1986, 103, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Rusnak, N.; Krisans, S.K. Diurnal variation of hmg-coa reductase activity in rat liver peroxisomes. Biochem. Biophys. Res. Commun. 1987, 148, 890–895. [Google Scholar] [CrossRef]
- Stamellos, K.D.; Shackelford, J.E.; Tanaka, R.D.; Krisans, S.K. Mevalonate kinase is localized in rat liver peroxisomes. J. Biol. Chem. 1992, 267, 5560–5568. [Google Scholar] [CrossRef]
- Stamellos, K.D.; Shackelford, J.E.; Shechter, I.; Jiang, G.; Conrad, D.; Keller, G.A.; Krisans, S.K. Subcellular localization of squalene synthase in rat hepatic cells. Biochemical and immunochemical evidence. J. Biol. Chem. 1993, 268, 12825–12836. [Google Scholar] [CrossRef]
- Krisans, S.K.; Ericsson, J.; Edwards, P.A.; Keller, G.A. Farnesyl-diphosphate synthase is localized in peroxisomes. J. Biol. Chem. 1994, 269, 14165–14169. [Google Scholar] [CrossRef]
- Faust, P.L.; Kovacs, W.J. Cholesterol biosynthesis and er stress in peroxisome deficiency. Biochimie 2014, 98, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, W.J.; Tape, K.N.; Shackelford, J.E.; Duan, X.; Kasumov, T.; Kelleher, J.K.; Brunengraber, H.; Krisans, S.K. Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem. Cell Biol. 2007, 127, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, W.J.; Olivier, L.M.; Krisans, S.K. Central role of peroxisomes in isoprenoid biosynthesis. Prog. Lipid Res. 2002, 41, 369–391. [Google Scholar] [CrossRef] [PubMed]
- Hogenboom, S.; Romeijn, G.J.; Houten, S.M.; Baes, M.; Wanders, R.J.; Waterham, H.R. Absence of functional peroxisomes does not lead to deficiency of enzymes involved in cholesterol biosynthesis. J. Lipid Res. 2002, 43, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Hogenboom, S.; Wanders, R.J.; Waterham, H.R. Cholesterol biosynthesis is not defective in peroxisome biogenesis defective fibroblasts. Mol. Genet. Metab. 2003, 80, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Hogenboom, S.; Tuyp, J.J.; Espeel, M.; Koster, J.; Wanders, R.J.; Waterham, H.R. Phosphomevalonate kinase is a cytosolic protein in humans. J. Lipid Res. 2004, 45, 697–705. [Google Scholar] [CrossRef]
- Hogenboom, S.; Tuyp, J.J.; Espeel, M.; Koster, J.; Wanders, R.J.; Waterham, H.R. Mevalonate kinase is a cytosolic enzyme in humans. J. Cell Sci. 2004, 117, 631–639. [Google Scholar] [CrossRef]
- Hogenboom, S.; Tuyp, J.J.; Espeel, M.; Koster, J.; Wanders, R.J.; Waterham, H.R. Human mevalonate pyrophosphate decarboxylase is localized in the cytosol. Mol. Genet. Metab. 2004, 81, 216–224. [Google Scholar] [CrossRef]
- Kovacs, W.J.; Charles, K.N.; Walter, K.M.; Shackelford, J.E.; Wikander, T.M.; Richards, M.J.; Fliesler, S.J.; Krisans, S.K.; Faust, P.L. Peroxisome deficiency-induced er stress and srebp-2 pathway activation in the liver of newborn pex2 knock-out mice. Biochim. Biophys. Acta 2012, 1821, 895–907. [Google Scholar] [CrossRef]
- Aboushadi, N.; Krisans, S.K. Analysis of isoprenoid biosynthesis in peroxisomal-deficient pex2 cho cell lines. J. Lipid Res. 1998, 39, 1781–1791. [Google Scholar] [CrossRef]
- Kovacs, W.J.; Shackelford, J.E.; Tape, K.N.; Richards, M.J.; Faust, P.L.; Fliesler, S.J.; Krisans, S.K. Disturbed cholesterol homeostasis in a peroxisome-deficient pex2 knockout mouse model. Mol Cell Biol 2004, 24, 1–13. [Google Scholar] [CrossRef]
- Charles, K.N.; Shackelford, J.E.; Faust, P.L.; Fliesler, S.J.; Stangl, H.; Kovacs, W.J. Functional peroxisomes are essential for efficient cholesterol sensing and synthesis. Front. Cell Dev. Biol. 2020, 8, 560266. [Google Scholar] [CrossRef]
- Chu, B.B.; Liao, Y.C.; Qi, W.; Xie, C.; Du, X.; Wang, J.; Yang, H.; Miao, H.H.; Li, B.L.; Song, B.L. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 2015, 161, 291–306. [Google Scholar] [CrossRef]
- Miyamoto, T.; Hosoba, K.; Itabashi, T.; Iwane, A.H.; Akutsu, S.N.; Ochiai, H.; Saito, Y.; Yamamoto, T.; Matsuura, S. Insufficiency of ciliary cholesterol in hereditary zellweger syndrome. EMBO J. 2020, 39, e103499. [Google Scholar] [CrossRef]
- Weinhofer, I.; Kunze, M.; Stangl, H.; Porter, F.D.; Berger, J. Peroxisomal cholesterol biosynthesis and smith-lemli-opitz syndrome. Biochem. Biophys. Res. Commun. 2006, 345, 205–209. [Google Scholar] [CrossRef]
- Buda, A.; Forss-Petter, S.; Hua, R.; Jaspers, Y.; Lassnig, M.; Waidhofer-Sollner, P.; Kemp, S.; Kim, P.; Weinhofer, I.; Berger, J. Abcd1 transporter deficiency results in altered cholesterol homeostasis. Biomolecules 2023, 13, 1333. [Google Scholar] [CrossRef]
- Weinhofer, I.; Forss-Petter, S.; Kunze, M.; Zigman, M.; Berger, J. X-linked adrenoleukodystrophy mice demonstrate abnormalities in cholesterol metabolism. FEBS Lett. 2005, 579, 5512–5516. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.L.; Menon, G.K.; Hanley, K.P. Hmg-coa reductase inhibitors perturb fatty acid metabolism and induce peroxisomes in keratinocytes. J. Lipid Res. 1992, 33, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Castro, I.G.; Schuldiner, M.; Zalckvar, E. Mind the organelle gap—Peroxisome contact sites in disease. Trends Biochem. Sci. 2018, 43, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. Acbd5 and vapb mediate membrane associations between peroxisomes and the er. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Hua, R.; Cheng, D.; Coyaud, E.; Freeman, S.; Di Pietro, E.; Wang, Y.; Vissa, A.; Yip, C.M.; Fairn, G.D.; Braverman, N.; et al. Vaps and acbd5 tether peroxisomes to the er for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 2017, 216, 367–377. [Google Scholar] [CrossRef]
- Chang, C.L.; Weigel, A.V.; Ioannou, M.S.; Pasolli, H.A.; Xu, C.S.; Peale, D.R.; Shtengel, G.; Freeman, M.; Hess, H.F.; Blackstone, C.; et al. Spastin tethers lipid droplets to peroxisomes and directs fatty acid trafficking through escrt-iii. J. Cell Biol. 2019, 218, 2583–2599. [Google Scholar] [CrossRef]
- Xiao, J.; Song, B.-L.; Luo, J. Peroxisomes in intracellular cholesterol transport: From basic physiology to brain pathology. Explor. Neuroprot. Ther. 2021, 1, 127–145. [Google Scholar] [CrossRef]
- Dorninger, F.; Brodde, A.; Braverman, N.E.; Moser, A.B.; Just, W.W.; Forss-Petter, S.; Brugger, B.; Berger, J. Homeostasis of phospholipids—The level of phosphatidylethanolamine tightly adapts to changes in ethanolamine plasmalogens. Biochim. Biophys. Acta 2015, 1851, 117–128. [Google Scholar] [CrossRef]
- Braverman, N.; Chen, L.; Lin, P.; Obie, C.; Steel, G.; Douglas, P.; Chakraborty, P.K.; Clarke, J.T.; Boneh, A.; Moser, A.; et al. Mutation analysis of pex7 in 60 probands with rhizomelic chondrodysplasia punctata and functional correlations of genotype with phenotype. Hum. Mutat. 2002, 20, 284–297. [Google Scholar] [CrossRef]
- Villoria-Gonzalez, A.; Zierfuss, B.; Parzer, P.; Heubock, E.; Zujovic, V.; Waidhofer-Sollner, P.; Ponleitner, M.; Rommer, P.; Gopfert, J.; Forss-Petter, S.; et al. Efficacy of hdac inhibitors in driving peroxisomal beta-oxidation and immune responses in human macrophages: Implications for neuroinflammatory disorders. Biomolecules 2023, 13, 1696. [Google Scholar] [CrossRef] [PubMed]
- de Virgilio, M.; Weninger, H.; Ivessa, N.E. Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J. Biol. Chem. 1998, 273, 9734–9743. [Google Scholar] [CrossRef] [PubMed]
- Wiesinger, C.; Kunze, M.; Regelsberger, G.; Forss-Petter, S.; Berger, J. Impaired very long-chain acyl-coa beta-oxidation in human x-linked adrenoleukodystrophy fibroblasts is a direct consequence of abcd1 transporter dysfunction. J. Biol. Chem. 2013, 288, 19269–19279. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M.; Neuberger, G.; Maurer-Stroh, S.; Ma, J.; Eck, T.; Braverman, N.; Schmid, J.A.; Eisenhaber, F.; Berger, J. Structural requirements for interaction of peroxisomal targeting signal 2 and its receptor pex7. J. Biol. Chem. 2011, 286, 45048–45062. [Google Scholar] [CrossRef] [PubMed]
- Yachnin, S.; Toub, D.B.; Mannickarottu, V. Divergence in cholesterol biosynthetic rates and 3-hydroxy-3-methylglutaryl-coa reductase activity as a consequence of granulocyte versus monocyte-macrophage differentiation in hl-60 cells. Proc. Natl. Acad. Sci. USA 1984, 81, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Breitling, R.; Krisans, S.K. A second gene for peroxisomal hmg-coa reductase? A genomic reassessment. J. Lipid Res. 2002, 43, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Engfelt, W.H.; Shackelford, J.E.; Aboushadi, N.; Jessani, N.; Masuda, K.; Paton, V.G.; Keller, G.A.; Krisans, S.K. Characterization of ut2 cells. The induction of peroxisomal 3-hydroxy-3-methylglutaryl-coenzyme a reductase. J. Biol. Chem. 1997, 272, 24579–24587. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.N.; Song, B.; DeBose-Boyd, R.A.; Ye, J. Sterol-regulated degradation of insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 2006, 281, 39308–39315. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M. The type-2 peroxisomal targeting signal. Biochim. Et Biophys. Acta. Mol. Cell Res. 2020, 1867, 118609. [Google Scholar] [CrossRef] [PubMed]
- Petriv, O.I.; Tang, L.; Titorenko, V.I.; Rachubinski, R.A. A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 2004, 341, 119–134. [Google Scholar] [CrossRef]
- Liscum, L.; Cummings, R.D.; Anderson, R.G.; DeMartino, G.N.; Goldstein, J.L.; Brown, M.S. 3-hydroxy-3-methylglutaryl-coa reductase: A transmembrane glycoprotein of the endoplasmic reticulum with n-linked “high-mannose” oligosaccharides. Proc. Natl. Acad. Sci. USA 1983, 80, 7165–7169. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Jimenez-Sanchez, G.; Koster, J.; Denis, S.; Van Roermund, C.W.; Silva-Zolezzi, I.; Moser, A.B.; Visser, W.F.; Gulluoglu, M.; Durmaz, O.; et al. A novel bile acid biosynthesis defect due to a deficiency of peroxisomal abcd3. Hum. Mol. Genet. 2015, 24, 361–370. [Google Scholar] [CrossRef]
- Biardi, L.; Sreedhar, A.; Zokaei, A.; Vartak, N.B.; Bozeat, R.L.; Shackelford, J.E.; Keller, G.A.; Krisans, S.K. Mevalonate kinase is predominantly localized in peroxisomes and is defective in patients with peroxisome deficiency disorders. J. Biol. Chem. 1994, 269, 1197–1205. [Google Scholar] [CrossRef]
| Cell Line | Cell Type | Differentiation/Stimulation | LDM Treatment | Lovastatin Treatment | Perc. Peroxisomal HMGCR (±STDEV) | Quantification |
|---|---|---|---|---|---|---|
| Monocyte | 7 d differentiation | 7 days | 16 h | 68 ± 8.7% | 147 (n = 3) | |
| THP-1 | 3 d differentiation | 3 days | 16 h | 50 ± 10.9% | 211 (n = 3) | |
| No differentiation | 3 days | 16 h | 14 ± 9.5% | 206 (n = 3) | ||
| U937 | Monocytic cell | 3 d differentiation | 3 days | 16 h | 0% | ND |
| HeLa | Epithelial cell | No differentiation | 3 days | 16 h | 0% | ND |
| HEK-293 | Embryonic kidney | No differentiation | 3 days | 16 h | 0% | ND |
| CHME-3 | Microglia cell | 1 d stimulation | 3 days | 16 h | <5% | ND |
| HepG2 | Liver cell | No differentiation | 3 days | 16 h | <3% | ND |
| Primary macrophage | 7 d differentiation | 3 days | 8 h * | 65 ± 8.8% | 320 (n = 3) | |
| Primary control fibroblast | No differentiation | 3 days | 16 h | 33 ± 10.0% | 201 (n = 3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Kunze, M.; Villoria-González, A.; Weinhofer, I.; Berger, J. Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions. Biomolecules 2024, 14, 244. https://doi.org/10.3390/biom14020244
Wang J, Kunze M, Villoria-González A, Weinhofer I, Berger J. Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions. Biomolecules. 2024; 14(2):244. https://doi.org/10.3390/biom14020244
Chicago/Turabian StyleWang, Jianqiu, Markus Kunze, Andrea Villoria-González, Isabelle Weinhofer, and Johannes Berger. 2024. "Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions" Biomolecules 14, no. 2: 244. https://doi.org/10.3390/biom14020244
APA StyleWang, J., Kunze, M., Villoria-González, A., Weinhofer, I., & Berger, J. (2024). Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions. Biomolecules, 14(2), 244. https://doi.org/10.3390/biom14020244








