Inflammation and Organic Cation Transporters Novel (OCTNs)
Abstract
1. Introduction
2. OCTN Functions and Dysfunctions
2.1. Relationships between Functions and Diseases
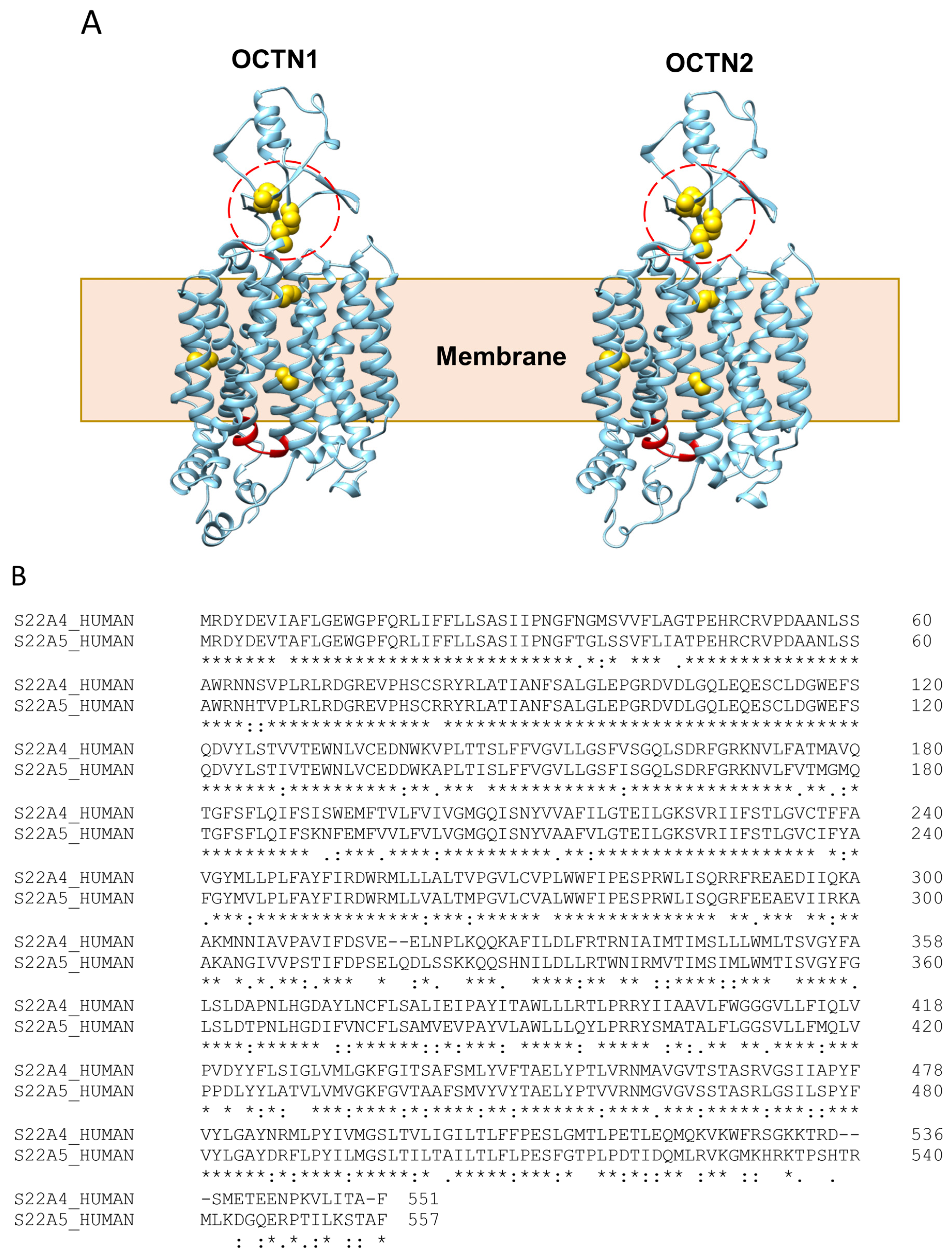
2.2. OCTN Polymorphisms and Relationships with Pathologies
3. OCTNs’ Role in Inflammation
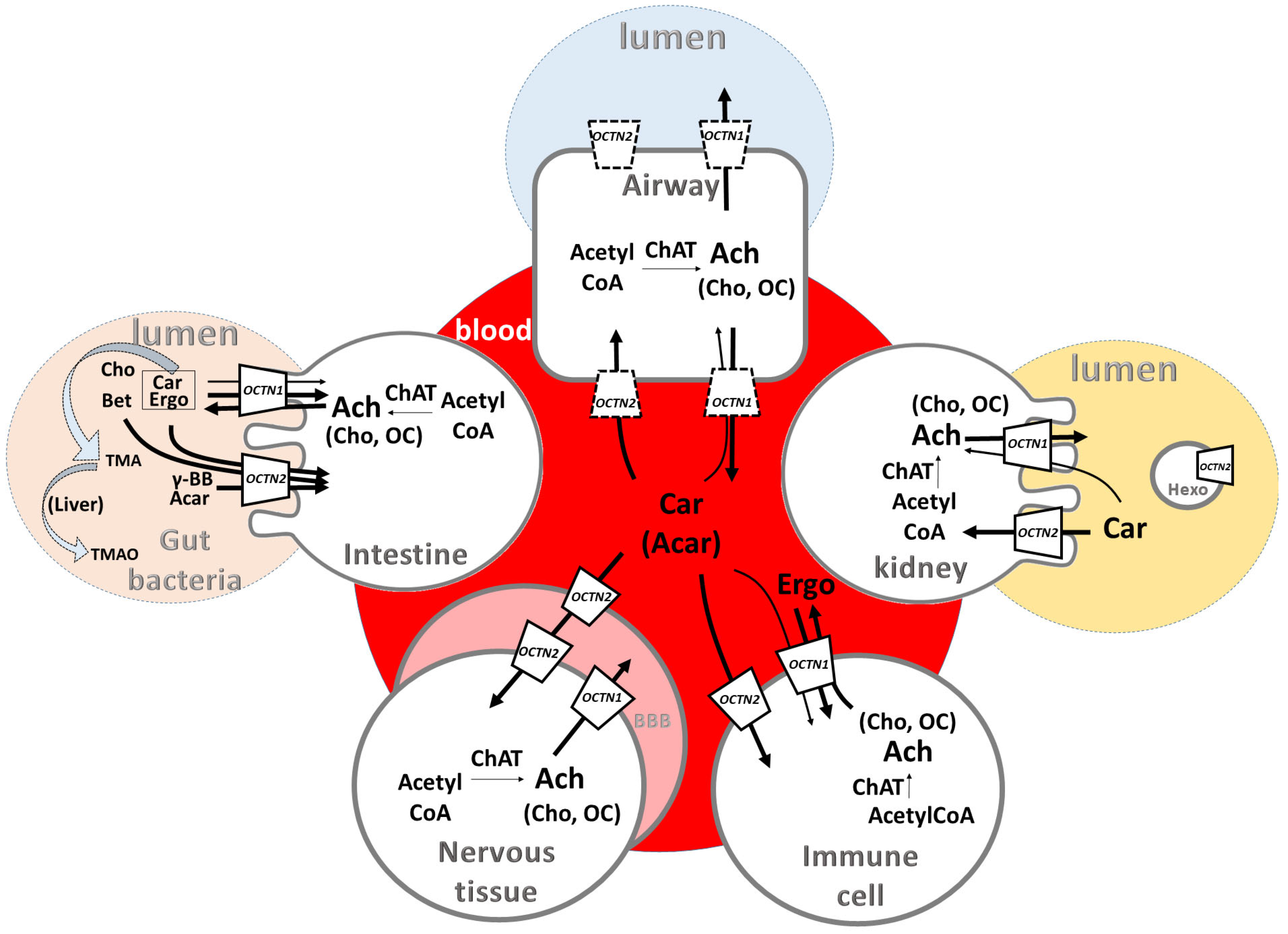
3.1. Involvement of OCTN Substrates in Inflammatory Processes
3.2. OCTN Substrates and Gut Microbiota Communication/Interconnections
4. Regulation of OCTN Expression and Inflammation
4.1. Major Players of OCTN Regulation
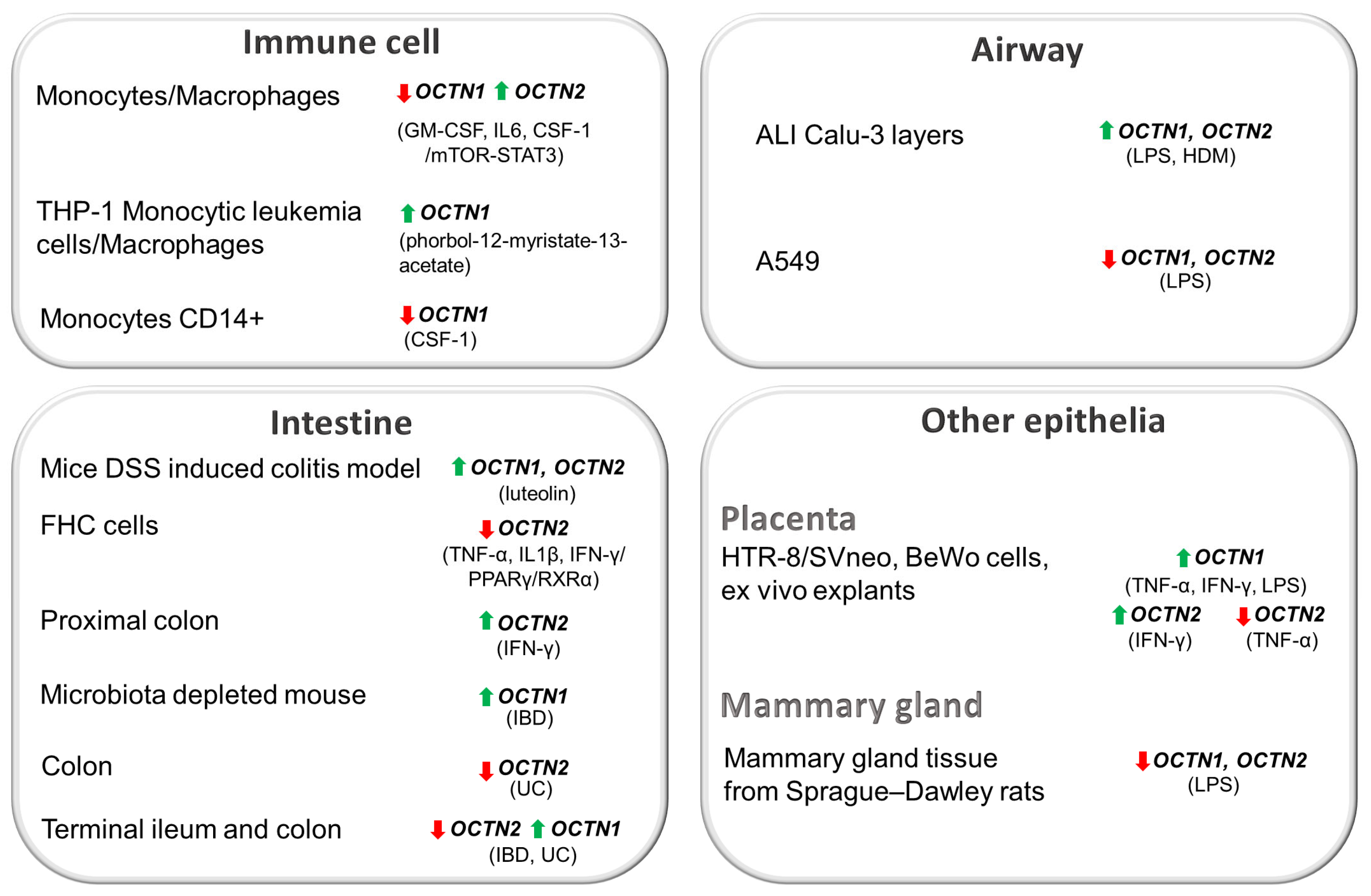
4.2. OCTN Regulation in Immune System
4.3. OCTN Regulation in Epithelia
4.3.1. Airway Epithelium
4.3.2. Gut Epithelium
4.3.3. Other Epithelia
5. Relationships of OCTNs with Altered Metabolite Profiles in Inflammation-Based Diseases
6. Perspectives and Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yu, W.; Wang, Z.; Yu, X.; Zhao, Y.; Xie, Z.; Zhang, K.; Chi, Z.; Chen, S.; Xu, T.; Jiang, D.; et al. Kir2.1-mediated membrane potential promotes nutrient acquisition and inflammation through regulation of nutrient transporters. Nat. Commun. 2022, 13, 3544. [Google Scholar] [CrossRef] [PubMed]
- Engelhart, D.C.; Granados, J.C.; Shi, D.; Saier, M.H., Jr.; Baker, M.E.; Abagyan, R.; Nigam, S.K. Systems Biology Analysis Reveals Eight SLC22 Transporter Subgroups, Including OATs, OCTs, and OCTNs. Int. J. Mol. Sci. 2020, 21, 1791. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Nigam, K.B.; Date, R.C.; Bush, K.T.; Springer, S.A.; Saier, M.H., Jr.; Wu, W.; Nigam, S.K. Evolutionary Analysis and Classification of OATs, OCTs, OCTNs, and Other SLC22 Transporters: Structure-Function Implications and Analysis of Sequence Motifs. PLoS ONE 2015, 10, e0140569. [Google Scholar] [CrossRef]
- Eraly, S.A.; Monte, J.C.; Nigam, S.K. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol. Genom. 2004, 18, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Galluccio, M.; Scalise, M.; Console, L.; Indiveri, C. OCTN: A Small Transporter Subfamily with Great Relevance to Human Pathophysiology, Drug Discovery, and Diagnostics. SLAS Discov. 2019, 24, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Koepsell, H. Organic Cation Transporters in Health and Disease. Pharmacol. Rev. 2020, 72, 253–319. [Google Scholar] [CrossRef] [PubMed]
- Masuo, Y.; Ohba, Y.; Yamada, K.; Al-Shammari, A.H.; Seba, N.; Nakamichi, N.; Ogihara, T.; Kunishima, M.; Kato, Y. Combination Metabolomics Approach for Identifying Endogenous Substrates of Carnitine/Organic Cation Transporter OCTN1. Pharm. Res. 2018, 35, 224. [Google Scholar] [CrossRef] [PubMed]
- Drenberg, C.D.; Gibson, A.A.; Pounds, S.B.; Shi, L.; Rhinehart, D.P.; Li, L.; Hu, S.; Du, G.; Nies, A.T.; Schwab, M.; et al. OCTN1 Is a High-Affinity Carrier of Nucleoside Analogues. Cancer Res. 2017, 77, 2102–2111. [Google Scholar] [CrossRef]
- Pochini, L.; Scalise, M.; Galluccio, M.; Pani, G.; Siminovitch, K.A.; Indiveri, C. The human OCTN1 (SLC22A4) reconstituted in liposomes catalyzes acetylcholine transport which is defective in the mutant L503F associated to the Crohn’s disease. Biochim. Biophys. Acta 2012, 1818, 559–565. [Google Scholar] [CrossRef]
- Pochini, L.; Scalise, M.; Di Silvestre, S.; Belviso, S.; Pandolfi, A.; Arduini, A.; Bonomini, M.; Indiveri, C. Acetylcholine and acetylcarnitine transport in peritoneum: Role of the SLC22A4 (OCTN1) transporter. Biochim. Biophys. Acta 2016, 1858, 653–660. [Google Scholar] [CrossRef]
- Pochini, L.; Scalise, M.; Indiveri, C. Immuno-detection of OCTN1 (SLC22A4) in HeLa cells and characterization of transport function. Int. Immunopharmacol. 2015, 29, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Grundemann, D.; Harlfinger, S.; Golz, S.; Geerts, A.; Lazar, A.; Berkels, R.; Jung, N.; Rubbert, A.; Schomig, E. Discovery of the ergothioneine transporter. Proc. Natl. Acad. Sci. USA 2005, 102, 5256–5261. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, M.; Nakamichi, N.; Yoshimura, T.; Masuo, Y.; Komori, T.; Ishimoto, T.; Matsuo, J.I.; Kato, Y. Homostachydrine is a Xenobiotic Substrate of OCTN1/SLC22A4 and Potentially Sensitizes Pentylenetetrazole-Induced Seizures in Mice. Neurochem. Res. 2020, 45, 2664–2678. [Google Scholar] [CrossRef] [PubMed]
- Juraszek, B.; Nalecz, K.A. SLC22A5 (OCTN2) Carnitine Transporter-Indispensable for Cell Metabolism, a Jekyll and Hyde of Human Cancer. Molecules 2020, 25, 14. [Google Scholar] [CrossRef]
- Ingoglia, F.; Visigalli, R.; Rotoli, B.M.; Barilli, A.; Riccardi, B.; Puccini, P.; Dall’Asta, V. Functional activity of L-carnitine transporters in human airway epithelial cells. Biochim. Biophys. Acta 2016, 1858, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Tamai, I.; Ohashi, R.; Nezu, J.I.; Sai, Y.; Kobayashi, D.; Oku, A.; Shimane, M.; Tsuji, A. Molecular and functional characterization of organic cation/carnitine transporter family in mice. J. Biol. Chem. 2000, 275, 40064–40072. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Barone, F.; Console, L.; Brunocilla, C.; Galluccio, M.; Scalise, M.; Indiveri, C. OCTN1 (SLC22A4) displays two different transport pathways for organic cations or zwitterions. Biochim. Biophys. Acta Biomembr. 2023, 1866, 184263. [Google Scholar] [CrossRef]
- Brockmoller, J.; Tzvetkov, M.V.; Hu, S. (Eds.) Organic Cation Transporter 1 (OCT1): Not Vital for Life, but of Substantial Biomedical Relevance. Front. Pharmacol. 2021, 11, 143. [Google Scholar]
- Nigam, S.K. The SLC22 Transporter Family: A Paradigm for the Impact of Drug Transporters on Metabolic Pathways, Signaling, and Disease. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 663–687. [Google Scholar] [CrossRef]
- Horvath, G.; Schmid, N.; Fragoso, M.A.; Schmid, A.; Conner, G.E.; Salathe, M.; Wanner, A. Epithelial organic cation transporters ensure pH-dependent drug absorption in the airway. Am. J. Respir. Cell Mol. Biol. 2007, 36, 53–60. [Google Scholar] [CrossRef]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Di Lascia, M.; Riccardi, B.; Puccini, P.; Dall’Asta, V.; Rotoli, B.M. Organic cation transporters (OCTs/OCTNs) in human primary alveolar epithelial cells. Biochem. Biophys. Res. Commun. 2021, 576, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.D.; Hu, X.; Ko, E.J.; Park, S.; Lee, Y.T.; Orr, M.; Fernandes, J.; Uppal, K.; Kang, S.M.; Jones, D.P.; et al. Metabolic pathways of lung inflammation revealed by high-resolution metabolomics (HRM) of H1N1 influenza virus infection in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R906–R916. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, K.; Catesson, A.; Griffin, J.L.; Holmes, E.; Williams, H.R.T. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J. Crohn’s Colitis 2021, 15, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.; Wright, N.J.; Guterres, H.; Fedor, J.G.; Butay, K.J.; Borgnia, M.J.; Im, W.; Lee, S.Y. Molecular basis of polyspecific drug binding and transport by OCT1 and OCT2. bioRxiv 2023. [Google Scholar] [CrossRef]
- Khanppnavar, B.; Maier, J.; Herborg, F.; Gradisch, R.; Lazzarin, E.; Luethi, D.; Yang, J.W.; Qi, C.; Holy, M.; Jantsch, K.; et al. Structural basis of organic cation transporter-3 inhibition. Nat. Commun. 2022, 13, 6714. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Scalise, M.; Galluccio, M.; Indiveri, C. OCTN cation transporters in health and disease: Role as drug targets and assay development. J. Biomol. Screen. 2013, 18, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Tamai, I. Pharmacological and pathophysiological roles of carnitine/organic cation transporters (OCTNs: SLC22A4, SLC22A5 and Slc22a21). Biopharm. Drug Dispos. 2013, 34, 29–44. [Google Scholar] [CrossRef]
- Wu, X.; Prasad, P.D.; Leibach, F.H.; Ganapathy, V. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem. Biophys. Res. Commun. 1998, 246, 589–595. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Tamai, I.; Yabuuchi, H.; Nezu, J.; Sai, Y.; Oku, A.; Shimane, M.; Tsuji, A. Cloning and characterization of a novel human pH-dependent organic cation transporter, OCTN1. FEBS Lett. 1997, 419, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Scalise, M.; Galluccio, M.; Amelio, L.; Indiveri, C. Reconstitution in liposomes of the functionally active human OCTN1 (SLC22A4) transporter overexpressed in Escherichia coli. Biochem. J. 2011, 439, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Pochini, L.; Peta, V.; Indiveri, C. Inhibition of the OCTN2 carnitine transporter by HgCl2 and methylmercury in the proteoliposome experimental model: Insights in the mechanism of toxicity. Toxicol. Mech. Methods 2013, 23, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Galluccio, M.; Pochini, L.; Peta, V.; Ianni, M.; Scalise, M.; Indiveri, C. Functional and molecular effects of mercury compounds on the human OCTN1 cation transporter: C50 and C136 are the targets for potent inhibition. Toxicol. Sci. 2015, 144, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Baba, S.P.; Bhatnagar, A. Role of Thiols in Oxidative Stress. Curr. Opin. Toxicol. 2018, 7, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Peltekova, V.D.; Wintle, R.F.; Rubin, L.A.; Amos, C.I.; Huang, Q.; Gu, X.; Newman, B.; Van Oene, M.; Cescon, D.; Greenberg, G.; et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat. Genet. 2004, 36, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Tokuhiro, S.; Yamada, R.; Chang, X.; Suzuki, A.; Kochi, Y.; Sawada, T.; Suzuki, M.; Nagasaki, M.; Ohtsuki, M.; Ono, M.; et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 2003, 35, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Selo, M.A.; Sake, J.A.; Ehrhardt, C.; Salomon, J.J. Organic Cation Transporters in the Lung-Current and Emerging (Patho)Physiological and Pharmacological Concepts. Int. J. Mol. Sci. 2020, 21, 9168. [Google Scholar] [CrossRef]
- Salomon, J.J.; Gausterer, J.C.; Selo, M.A.; Hosoya, K.I.; Huwer, H.; Schneider-Daum, N.; Lehr, C.M.; Ehrhardt, C. OCTN2-Mediated Acetyl-l-Carnitine Transport in Human Pulmonary Epithelial Cells In Vitro. Pharmaceutics 2019, 11, 396. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Saeterstad, S.; Ostvik, A.E.; Royset, E.S.; Bakke, I.; Sandvik, A.K.; Granlund, A.V.B. Profound gene expression changes in the epithelial monolayer of active ulcerative colitis and Crohn’s disease. PLoS ONE 2022, 17, e0265189. [Google Scholar] [CrossRef] [PubMed]
- Gopallawa, I.; Dehinwal, R.; Bhatia, V.; Gujar, V.; Chirmule, N. A four-part guide to lung immunology: Invasion, inflammation, immunity, and intervention. Front. Immunol. 2023, 14, 1119564. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Keulers, L.; Dehghani, A.; Knippels, L.; Garssen, J.; Papadopoulos, N.; Folkerts, G.; Braber, S.; van Bergenhenegouwen, J. Probiotics, prebiotics, and synbiotics to prevent or combat air pollution consequences: The gut-lung axis. Environ. Pollut. 2022, 302, 119066. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Martin, M.C.; Mendoza, J.L.; Taxonera, C.; Diaz-Rubio, M.; de la Concha, E.G.; Urcelay, E. Association of the organic cation transporter OCTN genes with Crohn’s disease in the Spanish population. Eur. J. Hum. Genet. 2006, 14, 222–226. [Google Scholar] [CrossRef]
- Defois, C.; Ratel, J.; Garrait, G.; Denis, S.; Le Goff, O.; Talvas, J.; Mosoni, P.; Engel, E.; Peyret, P. Food Chemicals Disrupt Human Gut Microbiota Activity And Impact Intestinal Homeostasis As Revealed By In Vitro Systems. Sci. Rep. 2018, 8, 11006. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nakamichi, N.; Nishijima, H.; Masuo, Y.; Kato, Y. Carnitine/Organic Cation Transporter OCTN1 Negatively Regulates Activation in Murine Cultured Microglial Cells. Neurochem. Res. 2018, 43, 116–128. [Google Scholar] [CrossRef]
- Waller, S.; Tremelling, M.; Bredin, F.; Godfrey, L.; Howson, J.; Parkes, M. Evidence for association of OCTN genes and IBD5 with ulcerative colitis. Gut 2006, 55, 809–814. [Google Scholar] [CrossRef][Green Version]
- Gulcin, I. Antioxidant and antiradical activities of L-carnitine. Life Sci. 2006, 78, 803–811. [Google Scholar] [CrossRef]
- Lee, B.J.; Lin, J.S.; Lin, Y.C.; Lin, P.T. Effects of L-carnitine supplementation on oxidative stress and antioxidant enzymes activities in patients with coronary artery disease: A randomized, placebo-controlled trial. Nutr. J. 2014, 13, 79. [Google Scholar] [CrossRef]
- Ribas, G.S.; Vargas, C.R.; Wajner, M. L-carnitine supplementation as a potential antioxidant therapy for inherited neurometabolic disorders. Gene 2014, 533, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Fortin, G.; Yurchenko, K.; Collette, C.; Rubio, M.; Villani, A.C.; Bitton, A.; Sarfati, M.; Franchimont, D. L-carnitine, a diet component and organic cation transporter OCTN ligand, displays immunosuppressive properties and abrogates intestinal inflammation. Clin. Exp. Immunol. 2009, 156, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Meng, F.; Li, H.; Hua, X.; Wu, L.; Yuan, X. L-carnitine alleviates oxidative stress-related damage via MAPK signaling in human lens epithelial cells exposed to H2O2. Int. J. Mol. Med. 2019, 44, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Grundemann, D. The ergothioneine transporter controls and indicates ergothioneine activity—A review. Prev. Med. 2012, 54, S71–S74. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yu, M.; Ochani, M.; Amella, C.A.; Tanovic, M.; Susarla, S.; Li, J.H.; Wang, H.; Yang, H.; Ulloa, L.; et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature 2003, 421, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.A.; Bassi, C.; Saunders, M.E.; Nechanitzky, R.; Morgado-Palacin, I.; Zheng, C.; Mak, T.W. Beyond neurotransmission: Acetylcholine in immunity and inflammation. J. Intern. Med. 2020, 287, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Halder, N.; Lal, G. Cholinergic System and Its Therapeutic Importance in Inflammation and Autoimmunity. Front. Immunol. 2021, 12, 660342. [Google Scholar] [CrossRef]
- Wessler, I.; Kirkpatrick, C.J. Acetylcholine beyond neurons: The non-neuronal cholinergic system in humans. Br. J. Pharmacol. 2008, 154, 1558–1571. [Google Scholar] [CrossRef]
- Kummer, W.; Krasteva-Christ, G. Non-neuronal cholinergic airway epithelium biology. Curr. Opin. Pharmacol. 2014, 16, 43–49. [Google Scholar] [CrossRef]
- Lips, K.S.; Volk, C.; Schmitt, B.M.; Pfeil, U.; Arndt, P.; Miska, D.; Ermert, L.; Kummer, W.; Koepsell, H. Polyspecific cation transporters mediate luminal release of acetylcholine from bronchial epithelium. Am. J. Respir. Cell Mol. Biol. 2005, 33, 79–88. [Google Scholar] [CrossRef]
- Nezu, J.; Tamai, I.; Oku, A.; Ohashi, R.; Yabuuchi, H.; Hashimoto, N.; Nikaido, H.; Sai, Y.; Koizumi, A.; Shoji, Y.; et al. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion-dependent carnitine transporter. Nat. Genet. 1999, 21, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.A.; DeLeeuw, S.; Coates, P.M.; Vianey-Liaud, C.; Divry, P.; Bonnefont, J.P.; Saudubray, J.M.; Haymond, M.; Trefz, F.K.; Breningstall, G.N.; et al. Chronic cardiomyopathy and weakness or acute coma in children with a defect in carnitine uptake. Ann. Neurol. 1991, 30, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, C.; Iacobazzi, V.; Tonazzi, A.; Giangregorio, N.; Infantino, V.; Convertini, P.; Console, L.; Palmieri, F. The mitochondrial carnitine/acylcarnitine carrier: Function, structure and physiopathology. Mol. Asp. Med. 2011, 32, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Grigat, S.; Fork, C.; Bach, M.; Golz, S.; Geerts, A.; Schomig, E.; Grundemann, D. The carnitine transporter SLC22A5 is not a general drug transporter, but it efficiently translocates mildronate. Drug Metab. Dispos. 2009, 37, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Kubo, Y.; Iwata, D.; Kato, S.; Sudo, T.; Sugiura, T.; Kagaya, T.; Wakayama, T.; Hirayama, A.; Sugimoto, M.; et al. Gene knockout and metabolome analysis of carnitine/organic cation transporter OCTN1. Pharm. Res. 2010, 27, 832–840. [Google Scholar] [CrossRef]
- Park, H.J.; Jung, E.S.; Kong, K.A.; Park, E.M.; Cheon, J.H.; Choi, J.H. Identification of OCTN2 variants and their association with phenotypes of Crohn’s disease in a Korean population. Sci. Rep. 2016, 6, 22887. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; Luo, J.; Zeng, Q.; Wang, M.; Bai, M.; Zhou, H.; Wang, J.; Jiang, H. Downregulation of OCTN2 by cytokines plays an important role in the progression of inflammatory bowel disease. Biochem. Pharmacol. 2020, 178, 114115. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Masuo, Y.; Takahashi, S.; Nakamichi, N.; Kato, Y. Organic cation transporter Octn1-mediated uptake of food-derived antioxidant ergothioneine into infiltrating macrophages during intestinal inflammation in mice. Drug Metab. Pharmacokinet. 2015, 30, 231–239. [Google Scholar] [CrossRef]
- Rock, R.B.; Gekker, G.; Aravalli, R.N.; Hu, S.; Sheng, W.S.; Peterson, P.K. Potentiation of HIV-1 expression in microglial cells by nicotine: Involvement of transforming growth factor-beta 1. J. Neuroimmune Pharmacol. 2008, 3, 143–149. [Google Scholar] [CrossRef]
- Martini, M.; Ferrara, A.M.; Giachelia, M.; Panieri, E.; Siminovitch, K.; Galeotti, T.; Larocca, L.M.; Pani, G. Association of the OCTN1/1672T variant with increased risk for colorectal cancer in young individuals and ulcerative colitis patients. Inflamm. Bowel Dis. 2012, 18, 439–448. [Google Scholar] [CrossRef]
- Haghighatdoost, F.; Jabbari, M.; Hariri, M. The effect of L-carnitine on inflammatory mediators: A systematic review and meta-analysis of randomized clinical trials. Eur. J. Clin. Pharmacol. 2019, 75, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, P.S.; Srinivas, S.R.; Matern, D.; Bennett, M.J.; Boriack, R.; George, V.; Xu, H.; Prasad, P.D.; Roon, P.; Ganapathy, V. Spontaneous development of intestinal and colonic atrophy and inflammation in the carnitine-deficient jvs (OCTN2(−/−)) mice. Mol. Genet. Metab. 2007, 92, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Sonne, S.; Shekhawat, P.S.; Matern, D.; Ganapathy, V.; Ignatowicz, L. Carnitine deficiency in OCTN2−/− newborn mice leads to a severe gut and immune phenotype with widespread atrophy, apoptosis and a pro-inflammatory response. PLoS ONE 2012, 7, e47729. [Google Scholar] [CrossRef] [PubMed]
- Ingoglia, F.; Visigalli, R.; Rotoli, B.M.; Barilli, A.; Riccardi, B.; Puccini, P.; Milioli, M.; Di Lascia, M.; Bernuzzi, G.; Dall’Asta, V. Human macrophage differentiation induces OCTN2-mediated L-carnitine transport through stimulation of mTOR-STAT3 axis. J. Leukoc. Biol. 2017, 101, 665–674. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, G.; Calvani, M.; Casamassimi, A.; Petillo, O.; Margarucci, S.; Rienzo, M.; Peluso, I.; Calvani, R.; Ciccodicola, A.; Caporaso, N.; et al. Experimental colitis: Decreased Octn2 and Atb0+ expression in rat colonocytes induces carnitine depletion that is reversible by carnitine-loaded liposomes. FASEB J. 2006, 20, 2544–2546. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.F.; Vachharajani, V.T.; Yoza, B.K.; McCall, C.E. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J. Biol. Chem. 2012, 287, 25758–25769. [Google Scholar] [CrossRef] [PubMed]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreno, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2019, 10, 2993. [Google Scholar] [CrossRef] [PubMed]
- Jamali-Raeufy, N.; Alizadeh, F.; Mehrabi, Z.; Mehrabi, S.; Goudarzi, M. Acetyl-L-carnitine confers neuroprotection against lipopolysaccharide (LPS) -induced neuroinflammation by targeting TLR4/NFkappaB, autophagy, inflammation and oxidative stress. Metab. Brain Dis. 2021, 36, 1391–1401. [Google Scholar] [CrossRef]
- Zahedi, E.; Sadr, S.S.; Sanaeierad, A.; Roghani, M. Chronic acetyl-L-carnitine treatment alleviates behavioral deficits and neuroinflammation through enhancing microbiota derived-SCFA in valproate model of autism. Biomed. Pharmacother. 2023, 163, 114848. [Google Scholar] [CrossRef]
- Miecz, D.; Januszewicz, E.; Czeredys, M.; Hinton, B.T.; Berezowski, V.; Cecchelli, R.; Nalecz, K.A. Localization of organic cation/carnitine transporter (OCTN2) in cells forming the blood-brain barrier. J. Neurochem. 2008, 104, 113–123. [Google Scholar] [CrossRef]
- Inano, A.; Sai, Y.; Nikaido, H.; Hasimoto, N.; Asano, M.; Tsuji, A.; Tamai, I. Acetyl-L-carnitine permeability across the blood-brain barrier and involvement of carnitine transporter OCTN2. Biopharm. Drug Dispos. 2003, 24, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Keshani, M.; Alikiaii, B.; Askari, G.; Yahyapoor, F.; Ferns, G.A.; Bagherniya, M. The effects of L-carnitine supplementation on inflammatory factors, oxidative stress, and clinical outcomes in patients with sepsis admitted to the intensive care unit (ICU): Study protocol for a double blind, randomized, placebo-controlled clinical trial. Trials 2022, 23, 170. [Google Scholar] [CrossRef] [PubMed]
- Yahyapoor, F.; Sedaghat, A.; Feizi, A.; Bagherniya, M.; Pahlavani, N.; Khadem-Rezaiyan, M.; Safarian, M.; Islam, M.S.; Zarifi, S.H.; Arabi, S.M.; et al. The effects of l-Carnitine supplementation on inflammatory markers, clinical status, and 28 days mortality in critically ill patients: A double-blind, randomized, placebo-controlled trial. Clin. Nutr. ESPEN 2022, 49, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, J.; Zheng, J.; Zhang, X.; Shao, J.; Zhao, L.; Hao, J. Anti-Inflammatory and Antioxidant Effects of Acetyl-L-Carnitine on Atherosclerotic Rats. Med. Sci. Monit. 2020, 26, e920250. [Google Scholar] [CrossRef] [PubMed]
- Mikhailova, T.L.; Sishkova, E.; Poniewierka, E.; Zhidkov, K.P.; Bakulin, I.G.; Kupcinskas, L.; Lesniakowski, K.; Grinevich, V.B.; Malecka-Panas, E.; Ardizzone, S.; et al. Randomised clinical trial: The efficacy and safety of propionyl-L-carnitine therapy in patients with ulcerative colitis receiving stable oral treatment. Aliment. Pharmacol. Ther. 2011, 34, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Ogawa, S.A.; Chau, L.; Whelan, K.A.; Hamilton, K.E.; Chen, J.; Tan, L.; Chen, E.Z.; Keilbaugh, S.; Fogt, F.; et al. Mitochondrial dysfunction in inflammatory bowel disease alters intestinal epithelial metabolism of hepatic acylcarnitines. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Lopez, E.; Zhong, Z.; Stubelius, A.; Sweeney, S.R.; Booshehri, L.M.; Antonucci, L.; Liu-Bryan, R.; Lodi, A.; Terkeltaub, R.; Lacal, J.C.; et al. Choline Uptake and Metabolism Modulate Macrophage IL-1beta and IL-18 Production. Cell Metab. 2019, 29, 1350–1362. [Google Scholar] [CrossRef] [PubMed]
- Judd, J.M.; Jasbi, P.; Winslow, W.; Serrano, G.E.; Beach, T.G.; Klein-Seetharaman, J.; Velazquez, R. Inflammation and the pathological progression of Alzheimer’s disease are associated with low circulating choline levels. Acta Neuropathol. 2023, 146, 565–583. [Google Scholar] [CrossRef]
- Wagner, C.A.; Lukewille, U.; Kaltenbach, S.; Moschen, I.; Broer, A.; Risler, T.; Broer, S.; Lang, F. Functional and pharmacological characterization of human Na(+)-carnitine cotransporter hOCTN2. Am. J. Physiol. Renal Physiol. 2000, 279, F584–F591. [Google Scholar] [CrossRef]
- Veskovic, M.; Mladenovic, D.; Milenkovic, M.; Tosic, J.; Borozan, S.; Gopcevic, K.; Labudovic-Borovic, M.; Dragutinovic, V.; Vucevic, D.; Jorgacevic, B.; et al. Betaine modulates oxidative stress, inflammation, apoptosis, autophagy, and Akt/mTOR signaling in methionine-choline deficiency-induced fatty liver disease. Eur. J. Pharmacol. 2019, 848, 39–48. [Google Scholar] [CrossRef]
- Zhou, S.; Xue, J.; Shan, J.; Hong, Y.; Zhu, W.; Nie, Z.; Zhang, Y.; Ji, N.; Luo, X.; Zhang, T.; et al. Gut-Flora-Dependent Metabolite Trimethylamine-N-Oxide Promotes Atherosclerosis-Associated Inflammation Responses by Indirect ROS Stimulation and Signaling Involving AMPK and SIRT1. Nutrients 2022, 14, 3338. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.C.; Hullar, M.A.J.; Randolph, T.W.; Franke, A.A.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Shepherd, J.A.; Madeleine, M.M.; Le Marchand, L.; et al. Associations of plasma trimethylamine N-oxide, choline, carnitine, and betaine with inflammatory and cardiometabolic risk biomarkers and the fecal microbiome in the Multiethnic Cohort Adiposity Phenotype Study. Am. J. Clin. Nutr. 2020, 111, 1226–1234. [Google Scholar] [CrossRef] [PubMed]
- Tacconi, E.; Palma, G.; De Biase, D.; Luciano, A.; Barbieri, M.; de Nigris, F.; Bruzzese, F. Microbiota Effect on Trimethylamine N-Oxide Production: From Cancer to Fitness-A Practical Preventing Recommendation and Therapies. Nutrients 2023, 15, 563. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, X.; Yang, F.; Zhao, R.; Pan, X.; Liang, J.; Tian, L.; Li, X.; Liu, L.; Xing, Y.; et al. Gut Microbiota-Dependent Marker TMAO in Promoting Cardiovascular Disease: Inflammation Mechanism, Clinical Prognostic, and Potential as a Therapeutic Target. Front. Pharmacol. 2019, 10, 1360. [Google Scholar] [CrossRef] [PubMed]
- Seldin, M.M.; Meng, Y.; Qi, H.; Zhu, W.; Wang, Z.; Hazen, S.L.; Lusis, A.J.; Shih, D.M. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-kappaB. J. Am. Heart Assoc. 2016, 5, e002767. [Google Scholar] [CrossRef] [PubMed]
- Constantino-Jonapa, L.A.; Espinoza-Palacios, Y.; Escalona-Montano, A.R.; Hernandez-Ruiz, P.; Amezcua-Guerra, L.M.; Amedei, A.; Aguirre-Garcia, M.M. Contribution of Trimethylamine N-Oxide (TMAO) to Chronic Inflammatory and Degenerative Diseases. Biomedicines 2023, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Vallance, H.D.; Koochin, A.; Branov, J.; Rosen-Heath, A.; Bosdet, T.; Wang, Z.; Hazen, S.L.; Horvath, G. Marked elevation in plasma trimethylamine-N-oxide (TMAO) in patients with mitochondrial disorders treated with oral l-carnitine. Mol. Genet. Metab. Rep. 2018, 15, 130–133. [Google Scholar] [CrossRef]
- Bader, S.; Diener, M. Segmental differences in the non-neuronal cholinergic system in rat caecum. Pflug. Arch. 2018, 470, 669–679. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, Y.; Guo, H.; Zhang, L.; Li, X.; Ringseis, R.; Wen, G.; Hui, D.; Liang, A.; Eder, K.; et al. Transcriptional regulation of the human, porcine and bovine OCTN2 gene by PPARalpha via a conserved PPRE located in intron 1. BMC Genet. 2014, 15, 90. [Google Scholar] [CrossRef][Green Version]
- Wada, E.; Koyanagi, S.; Kusunose, N.; Akamine, T.; Masui, H.; Hashimoto, H.; Matsunaga, N.; Ohdo, S. Modulation of peroxisome proliferator-activated receptor-alpha activity by bile acids causes circadian changes in the intestinal expression of Octn1/Slc22a4 in mice. Mol. Pharmacol. 2015, 87, 314–322. [Google Scholar] [CrossRef]
- Youssef, J.; Badr, M. Role of Peroxisome Proliferator-Activated Receptors in Inflammation Control. J. Biomed. Biotechnol. 2004, 2004, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Decara, J.; Rivera, P.; Lopez-Gambero, A.J.; Serrano, A.; Pavon, F.J.; Baixeras, E.; Rodriguez de Fonseca, F.; Suarez, J. Peroxisome Proliferator-Activated Receptors: Experimental Targeting for the Treatment of Inflammatory Bowel Diseases. Front. Pharmacol. 2020, 11, 730. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, G.; Petillo, O.; Margarucci, S.; Torpedine, A.; Calarco, A.; Koverech, A.; Boccia, A.; Paolella, G.; Peluso, G. Colon OCTN2 gene expression is up-regulated by peroxisome proliferator-activated receptor gamma in humans and mice and contributes to local and systemic carnitine homeostasis. J. Biol. Chem. 2010, 285, 27078–27087. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Ringseis, R.; Eder, K. Mouse OCTN2 is directly regulated by peroxisome proliferator-activated receptor alpha (PPARalpha) via a PPRE located in the first intron. Biochem. Pharmacol. 2010, 79, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Uray, I.P.; Mazumdar, A.; Mayer, J.A.; Brown, P.H. SLC22A5/OCTN2 expression in breast cancer is induced by estrogen via a novel intronic estrogen-response element (ERE). Breast Cancer Res. Treat. 2012, 134, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Qu, J.; Zhan, M.; Wu, L.X.; Zhang, Y.W.; Lou, X.Y.; Fu, L.J.; Zhou, H.H. Different involvement of promoter methylation in the expression of organic cation/carnitine transporter 2 (OCTN2) in cancer cell lines. PLoS ONE 2013, 8, e76474. [Google Scholar] [CrossRef] [PubMed]
- Buelow, D.R.; Anderson, J.T.; Pounds, S.B.; Shi, L.; Lamba, J.K.; Hu, S.; Gibson, A.A.; Goodwin, E.A.; Sparreboom, A.; Baker, S.D. DNA Methylation-Based Epigenetic Repression of SLC22A4 Promotes Resistance to Cytarabine in Acute Myeloid Leukemia. Clin. Transl. Sci. 2021, 14, 137–142. [Google Scholar] [CrossRef]
- Maeda, T.; Hirayama, M.; Kobayashi, D.; Miyazawa, K.; Tamai, I. Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis-associated transcriptional factor RUNX1 and inflammatory cytokines. Drug Metab. Dispos. 2007, 35, 394–401. [Google Scholar] [CrossRef]
- Karimian Pour, N.; McColl, E.R.; Piquette-Miller, M. Impact of Viral Inflammation on the Expression of Renal Drug Transporters in Pregnant Rats. Pharmaceutics 2019, 11, 624. [Google Scholar] [CrossRef]
- Harrach, S.; Edemir, B.; Schmidt-Lauber, C.; Pap, T.; Bertrand, J.; Ciarimboli, G. Importance of the novel organic cation transporter 1 for tyrosine kinase inhibition by saracatinib in rheumatoid arthritis synovial fibroblasts. Sci. Rep. 2017, 7, 1258. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ringseis, R.; Wen, G.; Eder, K. The pro-inflammatory cytokine tumor necrosis factor alpha stimulates expression of the carnitine transporter OCTN2 (novel organic cation transporter 2) and carnitine uptake via nuclear factor-kappaB in Madin-Darby bovine kidney cells. J. Dairy Sci. 2015, 98, 3840–3848. [Google Scholar] [CrossRef] [PubMed]
- Baillie, J.K.; Arner, E.; Daub, C.; De Hoon, M.; Itoh, M.; Kawaji, H.; Lassmann, T.; Carninci, P.; Forrest, A.R.; Hayashizaki, Y.; et al. Analysis of the human monocyte-derived macrophage transcriptome and response to lipopolysaccharide provides new insights into genetic aetiology of inflammatory bowel disease. PLoS Genet. 2017, 13, e1006641. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.; Cingolani, E.; Pritchard, D.I.; Bosquillon, C. Enhanced expression of Organic Cation Transporters in bronchial epithelial cell layers following insults associated with asthma—Impact on salbutamol transport. Eur. J. Pharm. Sci. 2017, 106, 62–70. [Google Scholar] [CrossRef]
- Rotoli, B.M.; Visigalli, R.; Barilli, A.; Ferrari, F.; Bianchi, M.G.; Di Lascia, M.; Riccardi, B.; Puccini, P.; Dall’Asta, V. Functional analysis of OCTN2 and ATB0,+ in normal human airway epithelial cells. PLoS ONE 2020, 15, e0228568. [Google Scholar] [CrossRef]
- Li, D.; Qi, C.; Zhou, J.; Wen, Z.; Zhu, X.; Xia, H.; Song, J. LPS-induced inflammation delays the transportation of ASP(+) due to down-regulation of OCTN1/2 in alveolar epithelial cells. J. Drug Target. 2020, 28, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Li, J.; Wu, B. Gene expression profiling of the mouse gut: Effect of intestinal flora on intestinal health. Mol. Med. Rep. 2018, 17, 3667–3673. [Google Scholar] [CrossRef]
- Fujiya, M.; Inaba, Y.; Musch, M.W.; Hu, S.; Kohgo, Y.; Chang, E.B. Cytokine regulation of OCTN2 expression and activity in small and large intestine. Inflamm. Bowel Dis. 2011, 17, 907–916. [Google Scholar] [CrossRef]
- Noble, C.L.; Abbas, A.R.; Cornelius, J.; Lees, C.W.; Ho, G.T.; Toy, K.; Modrusan, Z.; Pal, N.; Zhong, F.; Chalasani, S.; et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut 2008, 57, 1398–1405. [Google Scholar] [CrossRef]
- Wojtal, K.A.; Eloranta, J.J.; Hruz, P.; Gutmann, H.; Drewe, J.; Staumann, A.; Beglinger, C.; Fried, M.; Kullak-Ublick, G.A.; Vavricka, S.R. Changes in mRNA expression levels of solute carrier transporters in inflammatory bowel disease patients. Drug Metab. Dispos. 2009, 37, 1871–1877. [Google Scholar] [CrossRef]
- Palmieri, O.; Latiano, A.; Scimeca, D.; Bossa, F.; Corritore, G.; Latiano, T.; Andriulli, A.; Annese, V. IL23R, ATG16L1, IRGM, OCTN1, and OCTN2 mRNA expression in inflamed and noninflamed mucosa of IBD patients. Inflamm. Bowel Dis. 2011, 17, 1832–1833. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.; Alcorn, J. LPS-induced inflammation downregulates mammary gland glucose, fatty acid, and L-carnitine transporter expression at different lactation stages. Res. Vet. Sci. 2010, 89, 200–202. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; White, I.R.; Wilkinson, M.; Johnson, C.F.; Rattray, N.; Kishore, A.K.; Goodacre, R.; Smith, C.J.; Fowler, S.J. Breath and plasma metabolomics to assess inflammation in acute stroke. Sci. Rep. 2021, 11, 21949. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Li, R.; Antonov, A.A.; Wang, L.; Li, W.; Hua, Y.; Guo, H.; Wang, L.; Liu, P.; Chen, L.; et al. Discovery of Metabolite Biomarkers for Acute Ischemic Stroke Progression. J. Proteome Res. 2017, 16, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhou, K.; Li, H.; Dong, X.; Tan, G.; Chai, Y.; Wang, W.; Bi, X. Potential of serum metabolites for diagnosing post-stroke cognitive impairment. Mol. Biosyst. 2015, 11, 3287–3296. [Google Scholar] [CrossRef] [PubMed]
- Taubert, D.; Lazar, A.; Grimberg, G.; Jung, N.; Rubbert, A.; Delank, K.S.; Perniok, A.; Erdmann, E.; Schomig, E. Association of rheumatoid arthritis with ergothioneine levels in red blood cells: A case control study. J. Rheumatol. 2006, 33, 2139–2145. [Google Scholar] [PubMed]
- Coras, R.; Murillo-Saich, J.D.; Guma, M. Circulating Pro- and Anti-Inflammatory Metabolites and Its Potential Role in Rheumatoid Arthritis Pathogenesis. Cells 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.K.; Sharma, S.; Sharma, R.; Sinha, N.; Mandal, S.K.; Sharma, D. Metabolic fingerprinting of joint tissue of collagen-induced arthritis (CIA) rat: In vitro, high resolution NMR (nuclear magnetic resonance) spectroscopy based analysis. EXCLI J. 2018, 17, 257–272. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, T.; Lin, Y.; Xiong, M.; Chen, J.; Jian, C.; Zhang, J.; Xie, H.; Zeng, F.; Huang, Q.; et al. The change of plasma metabolic profile and gut microbiome dysbiosis in patients with rheumatoid arthritis. Front. Microbiol. 2022, 13, 931431. [Google Scholar] [CrossRef]
- Narasimhan, R.; Coras, R.; Rosenthal, S.B.; Sweeney, S.R.; Lodi, A.; Tiziani, S.; Boyle, D.; Kavanaugh, A.; Guma, M. Serum metabolomic profiling predicts synovial gene expression in rheumatoid arthritis. Arthritis Res. Ther. 2018, 20, 164. [Google Scholar] [CrossRef]
- Roivainen, A.; Parkkola, R.; Yli-Kerttula, T.; Lehikoinen, P.; Viljanen, T.; Mottonen, T.; Nuutila, P.; Minn, H. Use of positron emission tomography with methyl-11C-choline and 2-18F-fluoro-2-deoxy-D-glucose in comparison with magnetic resonance imaging for the assessment of inflammatory proliferation of synovium. Arthritis Rheum. 2003, 48, 3077–3084. [Google Scholar] [CrossRef] [PubMed]
- Hur, B.; Gupta, V.K.; Huang, H.; Wright, K.A.; Warrington, K.J.; Taneja, V.; Davis, J.M., 3rd; Sung, J. Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity. Arthritis Res. Ther. 2021, 23, 164. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, L.C.; Cole, J.; Rattigan, K.M.; Barrett, M.P.; Kurian, N.; McInnes, I.B.; Goodyear, C.S. The rheumatoid synovial environment alters fatty acid metabolism in human monocytes and enhances CCL20 secretion. Rheumatology 2020, 59, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Zeisbrich, M.; Yanes, R.E.; Zhang, H.; Watanabe, R.; Li, Y.; Brosig, L.; Hong, J.; Wallis, B.B.; Giacomini, J.C.; Assimes, T.L.; et al. Hypermetabolic macrophages in rheumatoid arthritis and coronary artery disease due to glycogen synthase kinase 3b inactivation. Ann. Rheum. Dis. 2018, 77, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Arra, M.; Abu-Amer, Y. Cross-talk of inflammation and chondrocyte intracellular metabolism in osteoarthritis. Osteoarthr. Cartil. 2023, 31, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Carlson, A.K.; Rawle, R.A.; Adams, E.; Greenwood, M.C.; Bothner, B.; June, R.K. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochem. Biophys. Res. Commun. 2018, 499, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Likhodii, S.; Zhang, Y.; Aref-Eshghi, E.; Harper, P.E.; Randell, E.; Green, R.; Martin, G.; Furey, A.; Sun, G.; et al. Classification of osteoarthritis phenotypes by metabolomics analysis. BMJ Open 2014, 4, e006286. [Google Scholar] [CrossRef]
- Mickiewicz, B.; Kelly, J.J.; Ludwig, T.E.; Weljie, A.M.; Wiley, J.P.; Schmidt, T.A.; Vogel, H.J. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. J. Orthop. Res. 2015, 33, 1631–1638. [Google Scholar] [CrossRef]
- Tootsi, K.; Kals, J.; Zilmer, M.; Paapstel, K.; Ottas, A.; Martson, A. Medium- and long-chain acylcarnitines are associated with osteoarthritis severity and arterial stiffness in end-stage osteoarthritis patients: A case-control study. Int. J. Rheum. Dis. 2018, 21, 1211–1218. [Google Scholar] [CrossRef]
- Li, M.; Yang, L.; Mu, C.; Sun, Y.; Gu, Y.; Chen, D.; Liu, T.; Cao, H. Gut microbial metabolome in inflammatory bowel disease: From association to therapeutic perspectives. Comput. Struct. Biotechnol. J. 2022, 20, 2402–2414. [Google Scholar] [CrossRef]
- Wu, X.; Liu, K.; Wu, Q.; Wang, M.; Chen, X.; Li, Y.; Qian, L.; Li, C.; Dai, G.; Zhang, Q.; et al. Biomarkers of Metabolomics in Inflammatory Bowel Disease and Damp-Heat Syndrome: A Preliminary Study. Evid. Based Complement. Altern. Med. 2022, 2022, 3319646. [Google Scholar] [CrossRef]
- Vich Vila, A.; Hu, S.; Andreu-Sanchez, S.; Collij, V.; Jansen, B.H.; Augustijn, H.E.; Bolte, L.A.; Ruigrok, R.; Abu-Ali, G.; Giallourakis, C.; et al. Faecal metabolome and its determinants in inflammatory bowel disease. Gut 2023, 72, 1472–1485. [Google Scholar] [CrossRef] [PubMed]
- Borren, N.Z.; Plichta, D.; Joshi, A.D.; Bonilla, G.; Sadreyev, R.; Vlamakis, H.; Xavier, R.J.; Ananthakrishnan, A.N. Multi-”-Omics” Profiling in Patients With Quiescent Inflammatory Bowel Disease Identifies Biomarkers Predicting Relapse. Inflamm. Bowel Dis. 2020, 26, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- Rattray, N.J.W.; Trivedi, D.K.; Xu, Y.; Chandola, T.; Johnson, C.H.; Marshall, A.D.; Mekli, K.; Rattray, Z.; Tampubolon, G.; Vanhoutte, B.; et al. Metabolic dysregulation in vitamin E and carnitine shuttle energy mechanisms associate with human frailty. Nat. Commun. 2019, 10, 5027. [Google Scholar] [CrossRef] [PubMed]
- Futatsugi, A.; Masuo, Y.; Kawabata, S.; Nakamichi, N.; Kato, Y. L503F variant of carnitine/organic cation transporter 1 efficiently transports metformin and other biguanides. J. Pharm. Pharmacol. 2016, 68, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Petito, V.; Fidaleo, M.; Pani, G.; Putignani, L.; Gasbarrini, A.; Scaldaferri, F. Tumor necrosis factor-alpha and solute carrier family 22 member 4 gene polymorphisms as potential determinants of intestinal dysbiosis. Dig. Liver. Dis. 2020, 52, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Song, G.G. Pathway analysis of a genome-wide association study of ileal Crohn’s disease. DNA Cell Biol. 2012, 31, 1549–1554. [Google Scholar] [CrossRef]
- Xuan, C.; Zhang, B.B.; Yang, T.; Deng, K.F.; Li, M.; Tian, R.J. Association between OCTN1/2 gene polymorphisms (1672C-T, 207G-C) and susceptibility of Crohn’s disease: A meta-analysis. Int. J. Colorectal. Dis. 2012, 27, 11–19. [Google Scholar] [CrossRef]
- Lin, Z.; Nelson, L.; Franke, A.; Poritz, L.; Li, T.Y.; Wu, R.; Wang, Y.; MacNeill, C.; Thomas, N.J.; Schreiber, S.; et al. OCTN1 variant L503F is associated with familial and sporadic inflammatory bowel disease. J. Crohns. Colitis. 2010, 4, 132–138. [Google Scholar] [CrossRef]
- Repnik, K.; Potocnik, U. Haplotype in the IBD5 region is associated with refractory Crohn’s disease in Slovenian patients and modulates expression of the SLC22A5 gene. J. Gastroenterol. 2011, 46, 1081–1091. [Google Scholar] [CrossRef]
- Angelini, S.; Pantaleo, M.A.; Ravegnini, G.; Zenesini, C.; Cavrini, G.; Nannini, M.; Fumagalli, E.; Palassini, E.; Saponara, M.; Di Battista, M.; et al. Polymorphisms in OCTN1 and OCTN2 transporters genes are associated with prolonged time to progression in unresectable gastrointestinal stromal tumours treated with imatinib therapy. Pharmacol. Res. 2013, 68, 1–6. [Google Scholar] [CrossRef]
- Ryckman, K.K.; Smith, C.J.; Jelliffe-Pawlowski, L.L.; Momany, A.M.; Berberich, S.L.; Murray, J.C. Metabolic heritability at birth: Implications for chronic disease research. Hum. Genet. 2014, 133, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Sim, W.H.; Ellis, J.A.; Ong, E.K.; Catto-Smith, A.G.; Cameron, D.J.; Bishop, R.F.; Kirkwood, C.D. Interaction of Crohn’s disease susceptibility genes in an Australian paediatric cohort. PLoS ONE 2010, 5, e15376. [Google Scholar] [CrossRef] [PubMed]
- Cucchiara, S.; Latiano, A.; Palmieri, O.; Staiano, A.M.; D’Inca, R.; Guariso, G.; Vieni, G.; Rutigliano, V.; Borrelli, O.; Valvano, M.R.; et al. Role of CARD15, DLG5 and OCTN genes polymorphisms in children with inflammatory bowel diseases. World J. Gastroenterol. 2007, 13, 1221–1229. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torkvist, L.; Noble, C.L.; Lordal, M.; Sjoqvist, U.; Lindforss, U.; Nimmo, E.R.; Lofberg, R.; Russell, R.K.; Satsangi, J. Contribution of the IBD5 locus to Crohn’s disease in the Swedish population. Scand. J. Gastroenterol. 2007, 42, 200–206. [Google Scholar] [CrossRef]
- Silverberg, M.S.; Duerr, R.H.; Brant, S.R.; Bromfield, G.; Datta, L.W.; Jani, N.; Kane, S.V.; Rotter, J.I.; Philip Schumm, L.; Hillary Steinhart, A.; et al. Refined genomic localization and ethnic differences observed for the IBD5 association with Crohn’s disease. Eur. J. Hum. Genet. 2007, 15, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolski, S.F.; McKinney, J.T.; Amat di San Filippo, C.; Giak Sim, K.; Wilcken, B.; Longo, N. Validation of dye-binding/high-resolution thermal denaturation for the identification of mutations in the SLC22A5 gene. Hum. Mutat. 2005, 25, 306–313. [Google Scholar] [CrossRef]
- Li, Y.; Chang, M.; Schrodi, S.J.; Callis-Duffin, K.P.; Matsunami, N.; Civello, D.; Bui, N.; Catanese, J.J.; Leppert, M.F.; Krueger, G.G.; et al. The 5q31 variants associated with psoriasis and Crohn’s disease are distinct. Hum. Mol. Genet. 2008, 17, 2978–2985. [Google Scholar] [CrossRef]
- Urban, T.J.; Gallagher, R.C.; Brown, C.; Castro, R.A.; Lagpacan, L.L.; Brett, C.M.; Taylor, T.R.; Carlson, E.J.; Ferrin, T.E.; Burchard, E.G.; et al. Functional genetic diversity in the high-affinity carnitine transporter OCTN2 (SLC22A5). Mol. Pharmacol. 2006, 70, 1602–1611. [Google Scholar] [CrossRef]
- Li, F.Y.; El-Hattab, A.W.; Bawle, E.V.; Boles, R.G.; Schmitt, E.S.; Scaglia, F.; Wong, L.J. Molecular spectrum of SLC22A5 (OCTN2) gene mutations detected in 143 subjects evaluated for systemic carnitine deficiency. Hum. Mutat. 2010, 31, E1632–E1651. [Google Scholar] [CrossRef]
- Lee, N.C.; Tang, N.L.; Chien, Y.H.; Chen, C.A.; Lin, S.J.; Chiu, P.C.; Huang, A.C.; Hwu, W.L. Diagnoses of newborns and mothers with carnitine uptake defects through newborn screening. Mol. Genet. Metab. 2010, 100, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Vaz, F.M.; Scholte, H.R.; Ruiter, J.; Hussaarts-Odijk, L.M.; Pereira, R.R.; Schweitzer, S.; de Klerk, J.B.; Waterham, H.R.; Wanders, R.J. Identification of two novel mutations in OCTN2 of three patients with systemic carnitine deficiency. Hum. Genet. 1999, 105, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Frigeni, M.; Balakrishnan, B.; Yin, X.; Calderon, F.R.O.; Mao, R.; Pasquali, M.; Longo, N. Functional and molecular studies in primary carnitine deficiency. Hum. Mutat. 2017, 38, 1684–1699. [Google Scholar] [CrossRef] [PubMed]
- Burwinkel, B.; Kreuder, J.; Schweitzer, S.; Vorgerd, M.; Gempel, K.; Gerbitz, K.D.; Kilimann, M.W. Carnitine transporter OCTN2 mutations in systemic primary carnitine deficiency: A novel Arg169Gln mutation and a recurrent Arg282ter mutation associated with an unconventional splicing abnormality. Biochem. Biophys. Res. Commun. 1999, 261, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Taroni, F.; Garavaglia, B.; Longo, N. Functional analysis of mutations in the OCTN2 transporter causing primary carnitine deficiency: Lack of genotype-phenotype correlation. Hum. Mutat. 2000, 16, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Korman, S.H.; Ye, J.; Gargus, J.J.; Gutman, A.; Taroni, F.; Garavaglia, B.; Longo, N. Phenotype and genotype variation in primary carnitine deficiency. Genet. Med. 2001, 3, 387–392. [Google Scholar] [CrossRef] [PubMed]
- El-Hattab, A.W.; Li, F.Y.; Shen, J.; Powell, B.R.; Bawle, E.V.; Adams, D.J.; Wahl, E.; Kobori, J.A.; Graham, B.; Scaglia, F.; et al. Maternal systemic primary carnitine deficiency uncovered by newborn screening: Clinical, biochemical, and molecular aspects. Genet. Med. 2010, 12, 19–24. [Google Scholar] [CrossRef]
- Schimmenti, L.A.; Crombez, E.A.; Schwahn, B.C.; Heese, B.A.; Wood, T.C.; Schroer, R.J.; Bentler, K.; Cederbaum, S.; Sarafoglou, K.; McCann, M.; et al. Expanded newborn screening identifies maternal primary carnitine deficiency. Mol. Genet. Metab. 2007, 90, 441–445. [Google Scholar] [CrossRef]
- Tang, M.F.; Sy, H.Y.; Kong, A.P.; Ko, F.W.; Wang, S.S.; Liu, T.C.; Chan, W.C.; Wong, G.W.; Hon, K.L.; Chan, J.C.; et al. Genetic effects of multiple asthma loci identified by genomewide association studies on asthma and spirometric indices. Pediatr. Allergy Immunol. 2016, 27, 185–194. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.C.; Kim, J.H.; Seo, Y.H.; Choi, S.J.; Ji, J.D.; Song, G.G. Meta-analysis of SLC22A4 and RUNX1 polymorphisms: Associations with rheumatoid arthritis susceptibility. Z. Rheumatol. 2015, 74, 351–358. [Google Scholar] [CrossRef]
- Ren, T.L.; Han, Z.J.; Yang, C.J.; Hang, Y.X.; Fang, D.Y.; Wang, K.; Zhu, X.; Ji, X.J.; Zhou, F.F. Association of SLC22A4 gene polymorphism with Rheumatoid arthritis in the Chinese population. J. Biochem. Mol. Toxicol. 2014, 28, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Cong, L.; Ionita-Laza, I.; Lo, S.H.; Zheng, T. Constructing gene association networks for rheumatoid arthritis using the backward genotype-trait association (BGTA) algorithm. BMC Proc. 2007, 1 (Suppl. S1), S13. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Song, J.J.; Kwon, D. Allelic based gene-gene interactions in rheumatoid arthritis. BMC Proc. 2009, 3 (Suppl. S7), S76. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Paradowska-Gorycka, A.; Safranow, K.; Dziedziejko, V.; Dutkiewicz, G.; Slucznowska-Glabowska, S.; Juzyszyn, Z.; Drozdzik, M. SLC22A5 polymorphism associated with risk of extra-articular manifestations in rheumatoid arthritis patients. Reumatologia 2019, 57, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Arimura, Y.; Saito, K.; Goto, A.; Motoya, S.; Shinomura, Y.; Miyamoto, A.; Imai, K. Association of SLC22A4/5 polymorphisms with steroid responsiveness of inflammatory bowel disease in Japan. Dis. Colon. Rectum. 2008, 51, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Zhang, G.; Zhang, F.; Ye, D.; Yang, D.; Yang, Y. Relationship Between SLC22A1 and SLC22A4 Gene Polymorphisms and Risk of Type 2 Diabetes in Chinese Han Population. Clin. Lab. 2018, 64, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R.K.; Zhou, L.; Nolte, I.M.; van der Steege, G.; van Dullemen, H.M.; Oosterom, E.; Bok, L.; Peppelenbosch, M.P.; Faber, K.N.; Kleibeuker, J.H.; et al. Runt-related transcription factor 3 is associated with ulcerative colitis and shows epistasis with solute carrier family 22, members 4 and 5. Inflamm. Bowel. Dis. 2008, 14, 1615–1622. [Google Scholar] [CrossRef]
- Yamase, Y.; Horibe, H.; Ueyama, C.; Fujimaki, T.; Oguri, M.; Kato, K.; Arai, M.; Watanabe, S.; Yamada, Y. Association of TOMM40 and SLC22A4 polymorphisms with ischemic stroke. Biomed. Rep. 2015, 3, 491–498. [Google Scholar] [CrossRef]
- Zou, D.; Lou, J.; Ke, J.; Mei, S.; Li, J.; Gong, Y.; Yang, Y.; Zhu, Y.; Tian, J.; Chang, J.; et al. Integrative expression quantitative trait locus-based analysis of colorectal cancer identified a functional polymorphism regulating SLC22A5 expression. Eur. J. Cancer 2018, 93, 1–9. [Google Scholar] [CrossRef]
- Sebastian-delaCruz, M.; Olazagoitia-Garmendia, A.; Gonzalez-Moro, I.; Santin, I.; Garcia-Etxebarria, K.; Castellanos-Rubio, A. Implication of m6A mRNA Methylation in Susceptibility to Inflammatory Bowel Disease. Epigenomes 2020, 4, 16. [Google Scholar] [CrossRef]
- Prieto-Perez, R.; Solano-Lopez, G.; Cabaleiro, T.; Roman, M.; Ochoa, D.; Talegon, M.; Baniandres, O.; Lopez-Estebaranz, J.L.; de la Cueva, P.; Dauden, E.; et al. Polymorphisms Associated with Age at Onset in Patients with Moderate-to-Severe Plaque Psoriasis. J. Immunol. Res. 2015, 2015, 101879. [Google Scholar] [CrossRef] [PubMed]
- de Ridder, L.; Weersma, R.K.; Dijkstra, G.; van der Steege, G.; Benninga, M.A.; Nolte, I.M.; Taminiau, J.A.; Hommes, D.W.; Stokkers, P.C. Genetic susceptibility has a more important role in pediatric-onset Crohn’s disease than in adult-onset Crohn’s disease. Inflamm. Bowel Dis. 2007, 13, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Rose, E.C.; di San Filippo, C.A.; Ndukwe Erlingsson, U.C.; Ardon, O.; Pasquali, M.; Longo, N. Genotype-phenotype correlation in primary carnitine deficiency. Hum. Mutat. 2012, 33, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, A.; Nozaki, J.; Ohura, T.; Kayo, T.; Wada, Y.; Nezu, J.; Ohashi, R.; Tamai, I.; Shoji, Y.; Takada, G.; et al. Genetic epidemiology of the carnitine transporter OCTN2 gene in a Japanese population and phenotypic characterization in Japanese pedigrees with primary systemic carnitine deficiency. Hum. Mol. Genet. 1999, 8, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Jaruskova, M.; Curik, N.; Hercog, R.; Polivkova, V.; Motlova, E.; Benes, V.; Klamova, H.; Pecherkova, P.; Belohlavkova, P.; Vrbacky, F.; et al. Genotypes of SLC22A4 and SLC22A5 regulatory loci are predictive of the response of chronic myeloid leukemia patients to imatinib treatment. J. Exp. Clin. Cancer Res. 2017, 36, 55. [Google Scholar] [CrossRef] [PubMed]
- Makhseed, N.; Vallance, H.D.; Potter, M.; Waters, P.J.; Wong, L.T.; Lillquist, Y.; Pasquali, M.; Amat di San Filippo, C.; Longo, N. Carnitine transporter defect due to a novel mutation in the SLC22A5 gene presenting with peripheral neuropathy. J. Inherit. Metab. Dis. 2004, 27, 778–780. [Google Scholar] [CrossRef] [PubMed]
- Mayatepek, E.; Nezu, J.; Tamai, I.; Oku, A.; Katsura, M.; Shimane, M.; Tsuji, A. Two novel missense mutations of the OCTN2 gene (W283R and V446F) in a patient with primary systemic carnitine deficiency. Hum. Mutat. 2000, 15, 118. [Google Scholar] [CrossRef]
- Ben Said, M.; Grati, M.; Ishimoto, T.; Zou, B.; Chakchouk, I.; Ma, Q.; Yao, Q.; Hammami, B.; Yan, D.; Mittal, R.; et al. A mutation in SLC22A4 encoding an organic cation transporter expressed in the cochlea strial endothelium causes human recessive non-syndromic hearing loss DFNB60. Hum. Genet. 2016, 135, 513–524. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pochini, L.; Galluccio, M.; Console, L.; Scalise, M.; Eberini, I.; Indiveri, C. Inflammation and Organic Cation Transporters Novel (OCTNs). Biomolecules 2024, 14, 392. https://doi.org/10.3390/biom14040392
Pochini L, Galluccio M, Console L, Scalise M, Eberini I, Indiveri C. Inflammation and Organic Cation Transporters Novel (OCTNs). Biomolecules. 2024; 14(4):392. https://doi.org/10.3390/biom14040392
Chicago/Turabian StylePochini, Lorena, Michele Galluccio, Lara Console, Mariafrancesca Scalise, Ivano Eberini, and Cesare Indiveri. 2024. "Inflammation and Organic Cation Transporters Novel (OCTNs)" Biomolecules 14, no. 4: 392. https://doi.org/10.3390/biom14040392
APA StylePochini, L., Galluccio, M., Console, L., Scalise, M., Eberini, I., & Indiveri, C. (2024). Inflammation and Organic Cation Transporters Novel (OCTNs). Biomolecules, 14(4), 392. https://doi.org/10.3390/biom14040392










