Biosynthesis of C4–C8 3-Hydroxycarboxylic Acids from Glucose through the Inverted Fatty Acid β-Oxidation by Metabolically Engineered Escherichia coli
Abstract
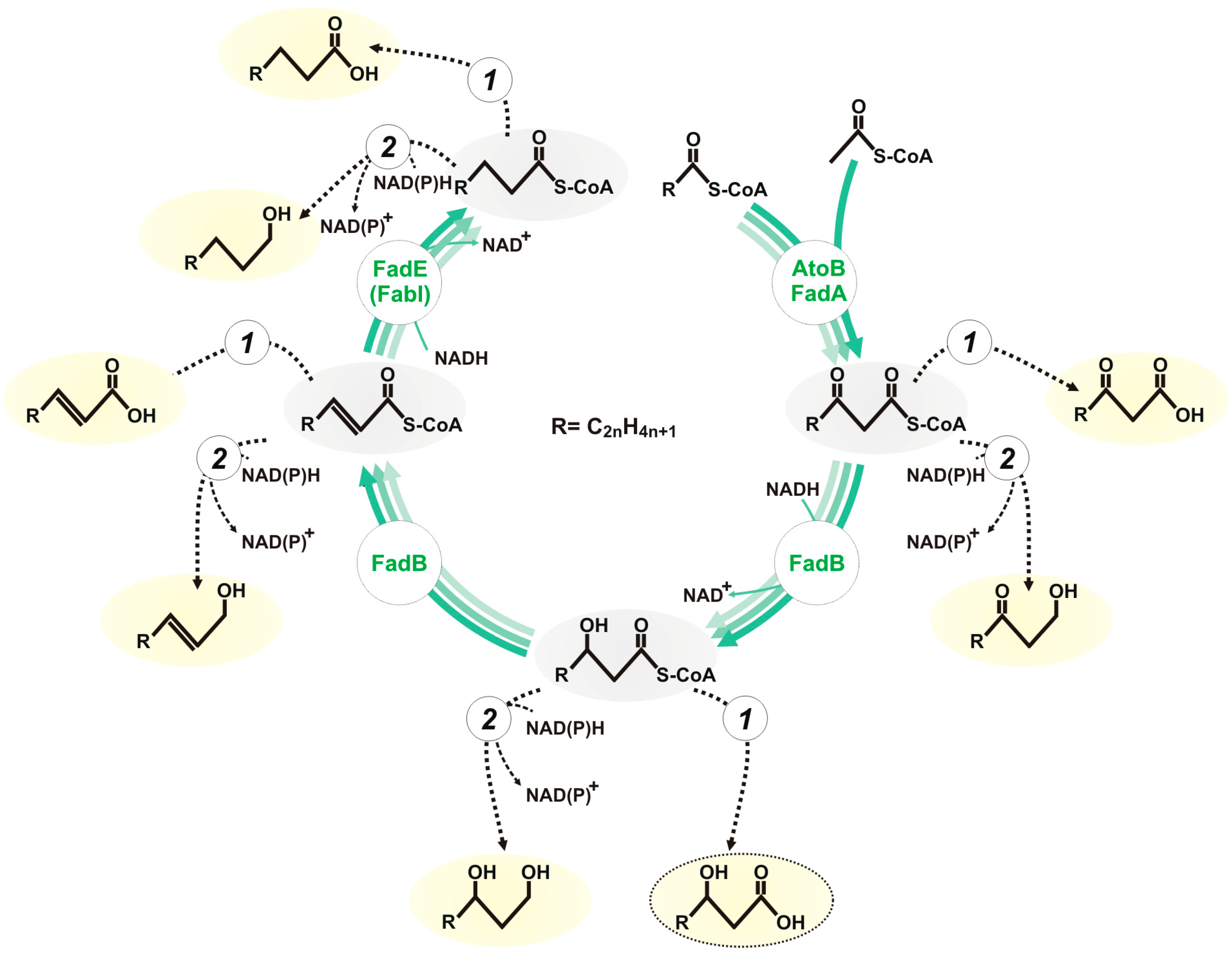
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Media
| Strain/Plasmid | Genotype | Reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | Wild-type E. coli K-12 strain VKPM B-6195 | VKPM |
| BOX3.3 Δ4 | MG1655 lacIQ, ∆ackA-pta, ∆poxB, ∆ldhA, ∆adhE, ∆fadE, PL-SDφ10-atoB, Ptrc-ideal-4-SDφ10-fadB, PL-SDφ10-tesB, ∆yciA | [20] |
| BOX3.3.1 | MG1655 lacIQ, ∆ackA-pta, ∆poxB, ∆ldhA, ∆adhE, PL-SDφ10-atoB, Ptrc-ideal-4-SDφ10-fadB, PL-SDφ10-tesB, ∆yciA, Ptrc-ideal-4-SDφ10-fadE | [9] |
| BOX3.3.2 | MG1655 lacIQ, ∆ackA-pta, ∆poxB, ∆ldhA, ∆adhE, PL-SDφ10-atoB, Ptrc-ideal-4-SDφ10-fadB, ∆fadE, PL-SDφ10-tesB, ∆yciA, Ptrc-ideal-4-SDφ10-fabI | [9] |
| BOX3.3.1.0 | BOX3.3.1 ∆tesA | This study |
| BOX3.3.2.0 | BOX3.3.2 ∆tesA | This study |
| BOX3.3.1.1 | BOX3.3.1 PL-SDφ10-fadB | This study |
| BOX3.3.2.1 | BOX3.3.2 PL-SDφ10-fadB | This study |
| BOX3.3.1.2 | BOX3.3.1 PL-SDφ10-fadB, PL-SDφ10-fadE | This study |
| BOX3.3.2.2 | BOX3.3.2 PL-SDφ10-fadB, PL-SDφ10-fabI | This study |
| BOX3.3.1.3 | BOX3.3.1 PL-SDφ10-fadB, PL-SDφ10-fadE, PL-SDφ10-fadA | This study |
| BOX3.3.2.3 | BOX3.3.2 PL-SDφ10-fadB, PL-SDφ10-fabI, PL-SDφ10-fadA | This study |
| Plasmids | ||
| pKD46 | pINT-ts, bla, ParaB-λgam-bet-exo | [26] |
| pMW118-(λattL-Cm-λattR) | pSC101, bla, cat, λattL-cat-λattR cassette | [27] |
| pMWts-Int/Xis | pSC101-ts, bla, PR-λxis-int, cIts857 | [28] |
2.2. Culturing of the Engineered Strains for Microaerobic Production of iBOX-Derived Compounds
2.3. Analytical Techniques
3. Results and Discussion
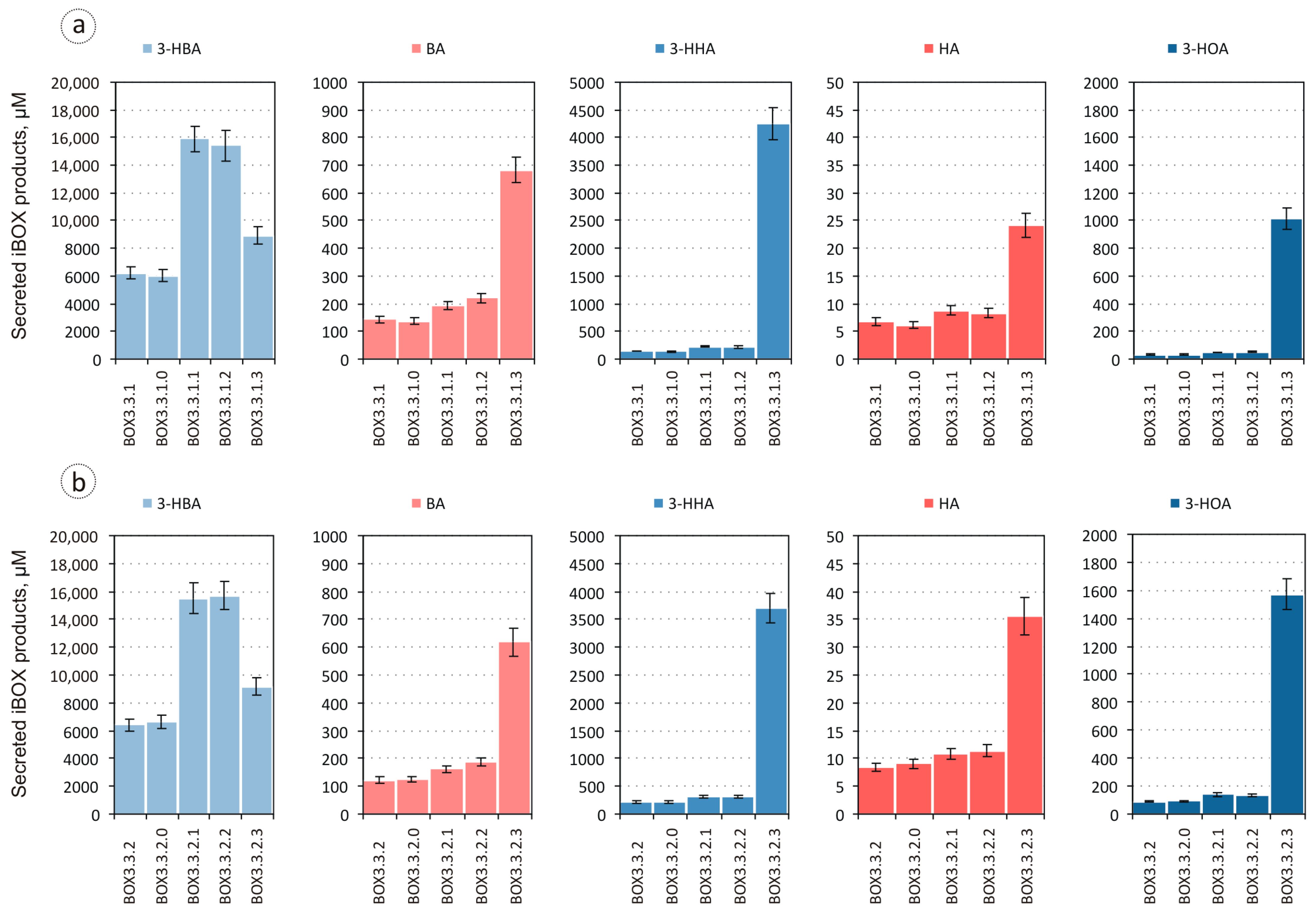
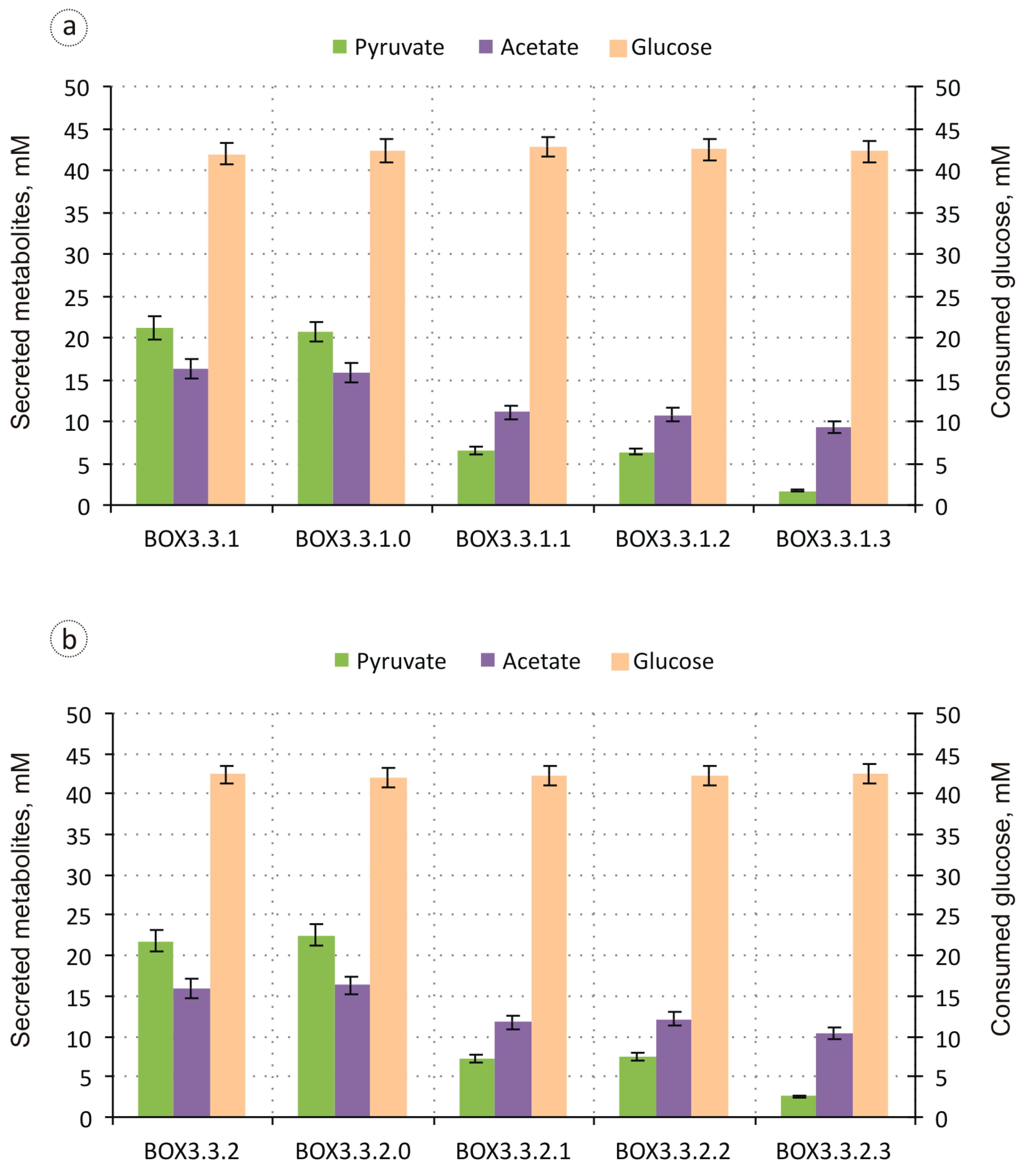
3.1. Evaluation of the Biosynthesis of 3-Hydroxycarboxylic Acids from Glucose through iBOX by Engineered E. coli Strains
3.2. Improvement in the Biosynthesis of 3-Hydroxycarboxylic Acids from Glucose through iBOX by Engineered E. coli Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pavoncello, V.; Barras, F.; Bouveret, E. Degradation of Exogenous Fatty Acids in Escherichia coli. Biomolecules 2022, 12, 1019. [Google Scholar] [CrossRef] [PubMed]
- Nandedkar, A.K.; Kumar, S. Biosynthesis of fatty acids in mammary tissue. II. Synthesis of butyrate in lactating rabbit mammary supernatant fraction by the reversal of beta-oxidation. Arch. Biochem. Biophys. 1969, 134, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Polen, T.; Bott, M.; Marienhagen, J. Reversal of β-oxidative pathways for the microbial production of chemicals and polymer building blocks. Metab. Eng. 2017, 42, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Tarasava, K.; Lee, S.H.; Chen, J.; Köpke, M.; Jewett, M.C.; Gonzalez, R. Reverse β-oxidation pathways for efficient chemical production. J. Ind. Microbiol. Biotechnol. 2022, 49, kuac003. [Google Scholar] [CrossRef] [PubMed]
- Dellomonaco, C.; Clomburg, J.M.; Miller, E.N.; Gonzalez, R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 2011, 476, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Sukhozhenko, A.V.; Shakulov, R.S.; Debabov, V.G. Metabolic engineering of Escherichia coli for 1-butanol biosynthesis through the inverted aerobic fatty acid β-oxidation pathway. Biotechnol. Lett. 2012, 34, 463–469. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Vick, J.E.; Blankschien, M.D.; Rodríguez-Moyá, M.; Gonzalez, R. A synthetic biology approach to engineer a functional reversal of the β-oxidation cycle. ACS Synth. Biol. 2012, 1, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Stasenko, A.A.; Shakulov, R.S.; Debabov, V.G. Metabolic engineering of Escherichia coli for 1,3-butanediol biosynthesis through the inverted fatty acid β-oxidation cycle. Appl. Biochem. Microbiol. 2016, 52, 15–22. [Google Scholar] [CrossRef]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Debabov, V.G. Evaluation of the efficiency of functional reversal of fatty acid β-oxidation in Escherichia coli upon the action of various native acyl-CoA dehydrogenases. Appl. Biochem. Microbiol. 2022, 58, 361–367. [Google Scholar] [CrossRef]
- Vick, J.E.; Clomburg, J.M.; Blankschien, M.D.; Chou, A.; Kim, S.; Gonzalez, R. Escherichia coli enoyl-acyl carrier protein reductase (FabI) supports efficient operation of a functional reversal of β-oxidation cycle. Appl. Environ. Microbiol. 2015, 81, 1406–1416. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, X.; Xia, X.; Dong, M. A systematic optimization of medium chain fatty acid biosynthesis via the reverse beta-oxidation cycle in Escherichia coli. Metab. Eng. 2017, 41, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Gonzalez, R. Selective production of decanoic acid from iterative reversal of β-oxidation pathway. Biotechnol. Bioeng. 2018, 115, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Clomburg, J.M.; Contreras, S.C.; Chou, A.; Siegel, J.B.; Gonzalez, R. Combination of type II fatty acid biosynthesis enzymes and thiolases supports a functional β-oxidation reversal. Metab. Eng. 2018, 45, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Mehrer, C.R.; Incha, M.R.; Politz, M.C.; Pfleger, B.F. Anaerobic production of medium-chain fatty alcohols via a β-reduction pathway. Metab. Eng. 2018, 48, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gonzalez, R. Engineering Escherichia coli for selective 1-decanol production using the reverse β-oxidation (rBOX) pathway. Metab. Eng. 2023, 79, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Clomburg, J.M.; Gonzalez, R. Synthesis of medium-chain length (C6-C10) fuels and chemicals via β-oxidation reversal in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2015, 42, 465–475. [Google Scholar] [CrossRef]
- Kim, S.; Cheong, S.; Gonzalez, R. Engineering Escherichia coli for the synthesis of short- and medium-chain α,β-unsaturated carboxylic acids. Metab. Eng. 2016, 36, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Q.; Wang, Q.; Liang, Q.; Qi, Q. Synthesis of polyhydroxyalkanoates from glucose that contain medium-chain-length monomers via the reversed fatty acid β-oxidation cycle in Escherichia coli. Metab. Eng. 2014, 24, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Sukhozhenko, A.V.; Debabov, V.G. Biosynthesis of enantiopure (S)-3-hydroxybutyrate from glucose through the inverted fatty acid β-oxidation pathway by metabolically engineered Escherichia coli. J. Biotechnol. 2017, 244, 16–24. [Google Scholar] [CrossRef]
- Gulevich, A.Y.; Skorokhodova, A.Y.; Debabov, V.G. Optimization of (S)-3-hydroxybutyric acid biosynthesis from glucose through the reversed fatty acid β-oxidation pathway by recombinant Escherichia coli strains. Appl. Biochem. Microbiol. 2021, 57, 161–169. [Google Scholar] [CrossRef]
- Blacklock, T.J.; Sohar, P.; Butcher, J.W.; Lamanec, T.; Grabowski, E.J.J. An enantioselective synthesis of the topically-active carbonic anhydrase inhibitor MK-0507: 5,6-dihydro-(S)-4-(ethylamino)-(S)-6-methyl-4H-thieno[2,3-b]thiopyran-2-sulfonamide 7,7-dioxide hydrochloride. J. Org. Chem. 1993, 58, 1672–1679. [Google Scholar] [CrossRef]
- Chiba, T.; Nakai, T. A synthetic approach to (+)-thienamycin from methylene (R)-3-hydroxybutanoate. A new entry to (3R,4R)-3-[(R)-1-hydroxyethyl]-4-acetoxy-2-azetidinone. Chem. Lett. 1985, 161, 651–654. [Google Scholar] [CrossRef]
- Mori, K. A simple synthesis of (S)-(+)-sulcatol, the pheromone of Gnathotrichus retusus, employing baker’s yeast for asymmetric reduction. Tetrahedron 1981, 37, 1341–1342. [Google Scholar] [CrossRef]
- Mori, K.; Takikawa, H. A new synthesis of the four stereoisomers of 3,11-dimethyl-2-nonacosanone, the female-produced sex pheromone of the german cockroach, Tetrahedron 1990, 46, 4473–4486. [CrossRef]
- Wang, Z.Q.; Song, H.; Koleski, E.J.; Hara, N.; Park, D.S.; Kumar, G.; Min, Y.; Dauenhauer, P.J.; Chang, M.C.Y. A dual cellular-heterogeneous catalyst strategy for the production of olefins from glucose. Nat. Chem. 2021, 13, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Katashkina, J.I.; Skorokhodova, A.Y.; Zimenkov, D.V.; Gulevich, A.Y.; Minaeva, N.I.; Doroshenko, V.G.; Biryukova, I.Y.; Mashko, S.V. Tuning the expression level of a gene located on a bacterial chromosome. Mol. Biol. 2005, 39, 719–726. [Google Scholar] [CrossRef]
- Katashkina, J.I.; Hara, Y.; Golubeva, L.I.; Andreeva, I.G.; Kuvaeva, T.M.; Mashko, S.V. Use of the λ Red-recombineering method for genetic engineering of Pantoea ananatis. BMC. Mol. Biol. 2009, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Manaiatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Veit, A.; Polen, T.; Wendisch, V.F. Global gene expression analysis of glucose overflow metabolism in Escherichia coli and reduction of aerobic acetate formation. Appl. Microbiol. Biotechnol. 2007, 74, 406–421. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 2005, 29, 765–794. [Google Scholar] [CrossRef]
- Bonner, W.M.; Bloch, K. Purification and properties of fatty acyl thioesterase I from Escherichia coli. J. Biol. Chem. 1972, 247, 3123–3133. [Google Scholar] [CrossRef]
- Nie, L.; Ren, Y.; Schulz, H. Identification and characterization of Escherichia coli thioesterase III that functions in fatty acid beta-oxidation. Biochemistry 2008, 47, 7744–7751. [Google Scholar] [CrossRef]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef] [PubMed]
- Camsund, D.; Heidorn, T.; Lindblad, P. Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng. 2014, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Deuschle, U.; Kammerer, W.; Gentz, R.; Bujard, H. Promoters of Escherichia coli: A hierarchy of in vivo strength indicates alternate structures. EMBO J. 1986, 5, 2987–2994. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.P.; Cronan, J.E. Two-carbon compounds and fatty acids as carbon sources. EcoSal Plus 2005, 1, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Binstock, J.F.; Schulz, H. Fatty acid oxidation complex from Escherichia coli. Methods Enzymol. 1981, 71, 403–411. [Google Scholar] [CrossRef]
- DiRusso, C.C. Primary sequence of the Escherichia coli fadBA operon, encoding the fatty acid-oxidizing multienzyme complex, indicates a high degree of homology to eucaryotic enzymes. J. Bacteriol. 1990, 172, 6459–6468. [Google Scholar] [CrossRef] [PubMed]
- Sah-Teli, S.; Hynönen, M.; Sulu, R.; Dalwani, S.; Schmitz, W.; Wierenga, R.; Venkatesan, R. Insights into the stability and substrate specificity of the E. coli aerobic beta-oxidation trifunctional enzyme complex. J. Struct. Biol. 2020, 210, 107494. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.N.; Lee, Y.; Hussein, R. Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Salis, H.M. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res. 2015, 43, 7137–7151. [Google Scholar] [CrossRef]
- Irastortza-Olaziregi, M.; Amster-Choder, O. Coupled transcription-translation in prokaryotes: An old couple with new surprises. Front. Microbiol. 2021, 11, 624830. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef] [PubMed]
- El-Mansi, M.; Cozzone, A.J.; Shiloach, J.; Eikmanns, B.J. Control of carbon flux through enzymes of central and intermediary metabolism during growth of Escherichia coli on acetate. Curr. Opin. Microbiol. 2006, 9, 173–179. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulevich, A.Y.; Skorokhodova, A.Y.; Debabov, V.G. Biosynthesis of C4–C8 3-Hydroxycarboxylic Acids from Glucose through the Inverted Fatty Acid β-Oxidation by Metabolically Engineered Escherichia coli. Biomolecules 2024, 14, 449. https://doi.org/10.3390/biom14040449
Gulevich AY, Skorokhodova AY, Debabov VG. Biosynthesis of C4–C8 3-Hydroxycarboxylic Acids from Glucose through the Inverted Fatty Acid β-Oxidation by Metabolically Engineered Escherichia coli. Biomolecules. 2024; 14(4):449. https://doi.org/10.3390/biom14040449
Chicago/Turabian StyleGulevich, Andrey Yu., Alexandra Yu. Skorokhodova, and Vladimir G. Debabov. 2024. "Biosynthesis of C4–C8 3-Hydroxycarboxylic Acids from Glucose through the Inverted Fatty Acid β-Oxidation by Metabolically Engineered Escherichia coli" Biomolecules 14, no. 4: 449. https://doi.org/10.3390/biom14040449





