Selection of Leptin Surrogates by a General Phenotypic Screening Method for Receptor Agonists
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Library Screening
2.3. Binding of the Antibody to hLepR on Live Cells
2.4. Western Blot for STAT Phosphorylation
2.5. Agonist Activity Determination by SIE-Luciferase/GFP Reporter System
2.6. Factor-Dependent Cell Proliferation Assay
2.7. Statistical Analyses
3. Results
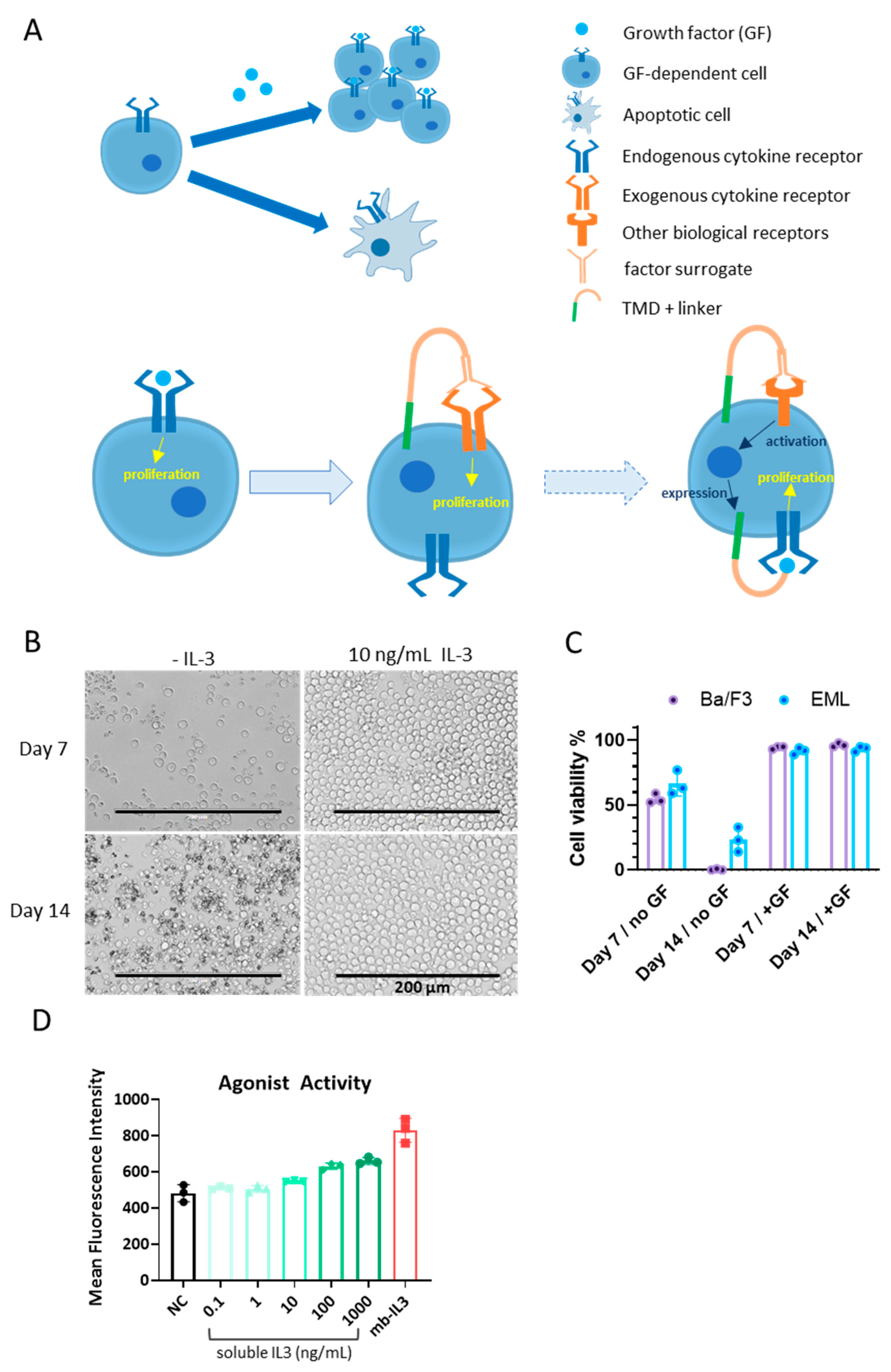
3.1. Cells for BRADS-Based Screening
3.2. Screening Based on the BRADS System
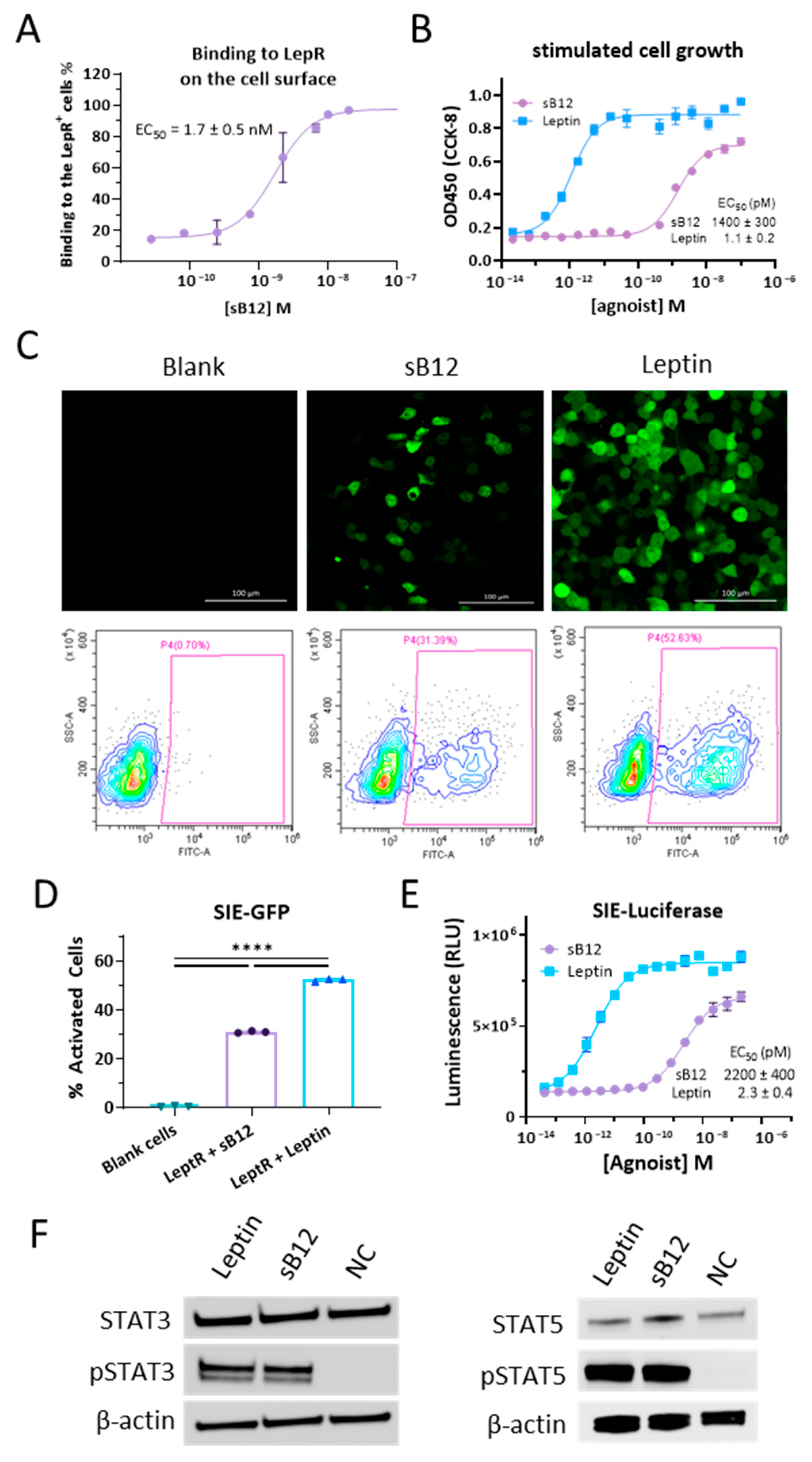
3.3. Characterizations of B12 Clone
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huse, W.D.; Sastry, L.; Iverson, S.A.; Kang, A.S.; Alting-Mees, M.; Burton, D.R.; Benkovic, S.J.; Lerner, R.A. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 1989, 246, 1275–1281. [Google Scholar] [CrossRef]
- Kang, A.S.; Barbas, C.F.; Janda, K.D.; Benkovic, S.J.; Lerner, R.A. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc. Natl. Acad. Sci. USA 1991, 88, 4363–4366. [Google Scholar] [CrossRef]
- Xu, L.; Carrer, A.; Zonta, F.; Qu, Z.; Ma, P.; Li, S.; Ceriani, F.; Buratto, D.; Crispino, G.; Zorzi, V.; et al. Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders. Front. Mol. Neurosci. 2017, 10, 298. [Google Scholar] [CrossRef]
- Lerner, R.A. Manufacturing immunity to disease in a test tube: The magic bullet realized. Angew. Chem. Int. Ed. Engl. 2006, 45, 8106–8125. [Google Scholar] [CrossRef]
- Qiang, M.; Dong, X.; Zha, Z.; Zuo, X.K.; Song, X.L.; Zhao, L.; Yuan, C.; Huang, C.; Tao, P.; Hu, Q.; et al. Selection of an ASIC1a-blocking combinatorial antibody that protects cells from ischemic death. Proc. Natl. Acad. Sci. USA 2018, 115, E7469–E7477. [Google Scholar] [CrossRef]
- Shi, X.; Wan, Y.; Wang, N.; Xiang, J.; Wang, T.; Yang, X.; Wang, J.; Dong, X.; Dong, L.; Yan, L.; et al. Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms. Nat. Commun. 2021, 12, 2547. [Google Scholar] [CrossRef]
- Li, W.; Wang, F.; Li, Y.; Yan, L.; Liu, L.; Zhu, W.; Ma, P.; Shi, X.; Yang, G. Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System. Vaccines 2023, 11, 771. [Google Scholar] [CrossRef]
- Kuang, Y.; Zorzi, V.; Buratto, D.; Ziraldo, G.; Mazzarda, F.; Peres, C.; Nardin, C.; Salvatore, A.M.; Chiani, F.; Scavizzi, F.; et al. A potent antagonist antibody targeting connexin hemichannels alleviates Clouston syndrome symptoms in mutant mice. EBioMedicine 2020, 57, 102825. [Google Scholar] [CrossRef]
- Kroeze, W.K.; Sassano, M.F.; Huang, X.P.; Lansu, K.; McCorvy, J.D.; Giguere, P.M.; Sciaky, N.; Roth, B.L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22, 362–369. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, H.; Yea, K.; Lerner, R.A. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 8099–8104. [Google Scholar] [CrossRef]
- Lerner, R.A. Combinatorial antibody libraries: New advances, new immunological insights. Nat. Rev. Immunol. 2016, 16, 98–508. [Google Scholar] [CrossRef]
- Stepanov, A.V.; Markov, O.V.; Chernikov, I.V.; Gladkikh, D.V.; Zhang, H.; Jones, T.; Sen’kova, A.V.; Chernolovskaya, E.L.; Zenkova, M.A.; Kalinin, R.S.; et al. Autocrine-based selection of ligands for personalized CAR-T therapy of lymphoma. Sci. Adv. 2018, 4, eaau4580. [Google Scholar] [CrossRef]
- Lerner, R.A.; Grover, R.K.; Zhang, H.; Xie, J.; Han, K.H.; Peng, Y.; Yea, K. Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q. Rev. Biophys. 2015, 48, 389–394. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Xie, J.; El-Mecharrafie, N.; Gross, S.; Lee, S.; Lerner, R.A.; Baldwin, K.K. Replacing reprogramming factors with antibodies selected from combinatorial antibody libraries. Nat. Biotechnol. 2017, 35, 960–968. [Google Scholar] [CrossRef]
- Caron, A.; Lee, S.; Elmquist, J.K.; Gautron, L. Leptin and brain-adipose crosstalks. Nat. Rev. Neurosci. 2018, 19, 153–165. [Google Scholar] [CrossRef]
- Xu, J.; Bartolome, C.L.; Low, C.S.; Yi, X.; Chien, C.H.; Wang, P.; Kong, D. Genetic identification of leptin neural circuits in energy and glucose homeostases. Nature 2018, 556, 505–509. [Google Scholar] [CrossRef]
- Zeng, W.; Pirzgalska, R.M.; Pereira, M.M.; Kubasova, N.; Barateiro, A.; Seixas, E.; Lu, Y.H.; Kozlova, A.; Voss, H.; Martins, G.G.; et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 2015, 163, 84–94. [Google Scholar] [CrossRef]
- Karsenty, G.; Khosla, S. The crosstalk between bone remodeling and energy metabolism: A translational perspective. Cell Metab. 2022, 34, 805–817. [Google Scholar] [CrossRef]
- Briffa, J.F.; McAinch, A.J.; Romano, T.; Wlodek, M.E.; Hryciw, D.H. Leptin in pregnancy and development: A contributor to adulthood disease? Am. J. Physiol. Endocrinol. Metab. 2015, 308, E335–E350. [Google Scholar] [CrossRef]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; Ordovas, J.M.; Cupples, L.A.; Defoort, C.; Lovegrove, J.A.; Drevon, C.A.; Blaak, E.E.; et al. Leptin receptor polymorphisms interact with polyunsaturated fatty acids to augment risk of insulin resistance and metabolic syndrome in adults. J. Nutr. 2010, 140, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Cassano, S.; Pucino, V.; La Rocca, C.; Procaccini, C.; De Rosa, V.; Marone, G.; Matarese, G. Leptin modulates autophagy in human CD4+CD25- conventional T cells. Metabolism 2014, 63, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Kuang, Y.; Li, Y.; Li, W.; Gao, Z.; Liu, L.; Qiang, M.; Zha, Z.; Fan, K.; Ma, P.; et al. Selection of a Full Agonist Combinatorial Antibody that Rescues Leptin Deficiency In Vivo. Adv. Sci. 2020, 7, 2000818. [Google Scholar] [CrossRef]
- Altarejos, J.Y.; Pangilinan, J.; Podgrabinska, S.; Akinci, B.; Foss-Freitas, M.; Neidert, A.H.; Ray, Y.; Zheng, W.J.; Kim, S.; Kamat, V.; et al. Preclinical, randomized phase 1, and compassionate use evaluation of REGN4461, a leptin receptor agonist antibody for leptin deficiency. Sci. Transl. Med. 2023, 15, eadd4897. [Google Scholar] [CrossRef]
- Charrier, S.; Ferrand, M.; Zerbato, M.; Precigout, G.; Viornery, A.; Bucher-Laurent, S.; Benkhelifa-Ziyyat, S.; Merten, O.W.; Perea, J.; Galy, A. Quantification of lentiviral vector copy numbers in individual hematopoietic colony-forming cells shows vector dose-dependent effects on the frequency and level of transduction. Gene Ther. 2011, 18, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Imamura, M.; Shook, D.; Kamiya, T.; Shimasaki, N.; Chai, S.M.; Coustan-Smith, E.; Imai, C.; Campana, D. Autonomous growth and increased cytotoxicity of natural killer cells expressing membrane-bound interleukin-15. Blood 2014, 124, 1081–1088. [Google Scholar] [CrossRef]
- Lang, R.A.; Metcalf, D.; Gough, N.M.; Dunn, A.R.; Gonda, T.J. Expression of a hemopoietic growth factor cDNA in a factor-dependent cell line results in autonomous growth and tumorigenicity. Cell 1985, 43, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Choong, M.L.; Yong, Y.P.; Tan, A.C.; Luo, B.; Lodish, H.F. LIX: A chemokine with a role in hematopoietic stem cells maintenance. Cytokine 2004, 25, 239–245. [Google Scholar] [CrossRef]
- Ye, Z.J.; Kluger, Y.; Lian, Z.; Weissman, S.M. Two types of precursor cells in a multipotential hematopoietic cell line. Proc. Natl. Acad. Sci. USA 2005, 102, 18461–18466. [Google Scholar] [CrossRef]
- Lee, H.M.; Zhang, H.; Schulz, V.; Tuck, D.P.; Forget, B.G. Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells. Blood 2010, 116, 720–730. [Google Scholar] [CrossRef]
- Ye, Z.J.; Gulcicek, E.; Stone, K.; Lam, T.; Schulz, V.; Weissman, S.M. Complex interactions in EML cell stimulation by stem cell factor and IL-3. Proc. Natl. Acad. Sci. USA 2011, 108, 4882–4887. [Google Scholar] [CrossRef]
- Wu, J.Q.; Seay, M.; Schulz, V.P.; Hariharan, M.; Tuck, D.; Lian, J.; Du, J.; Shi, M.; Ye, Z.; Gerstein, M.; et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS Genet. 2012, 8, e1002565. [Google Scholar] [CrossRef] [PubMed]
- Greco, M.; De Santo, M.; Comandè, A.; Belsito, E.L.; Andò, S.; Liguori, A.; Leggio, A. Leptin-Activity Modulators and Their Potential Pharmaceutical Applications. Biomolecules 2021, 11, 1045. [Google Scholar] [CrossRef]
- Morsut, L.; Roybal, K.T.; Xiong, X.; Gordley, R.M.; Coyle, S.M.; Thomson, M.; Lim, W.A. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 2016, 164, 780–791. [Google Scholar] [CrossRef]
- Neri, D.; Lerner, R.A. DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem. 2018, 87, 479–502. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Chen, X.; Yang, G.; Shi, X. Selection of Leptin Surrogates by a General Phenotypic Screening Method for Receptor Agonists. Biomolecules 2024, 14, 457. https://doi.org/10.3390/biom14040457
Wang T, Chen X, Yang G, Shi X. Selection of Leptin Surrogates by a General Phenotypic Screening Method for Receptor Agonists. Biomolecules. 2024; 14(4):457. https://doi.org/10.3390/biom14040457
Chicago/Turabian StyleWang, Tao, Xixi Chen, Guang Yang, and Xiaojie Shi. 2024. "Selection of Leptin Surrogates by a General Phenotypic Screening Method for Receptor Agonists" Biomolecules 14, no. 4: 457. https://doi.org/10.3390/biom14040457





