Hyaluronic Acid Prevents Fusion of Brain Tumor-Derived Spheroids and Selectively Alters Their Gene Expression Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Spheroids
2.3. Spheroid Viability Assays
2.4. Gene Expression Analysis
2.5. Statistical Analysis
3. Results
3.1. Cell Cultures Derived from Tissue Strains of Brain Tumors
3.2. Spheroids Derived from Cell Cultures
3.3. Spheroid Viability Assays
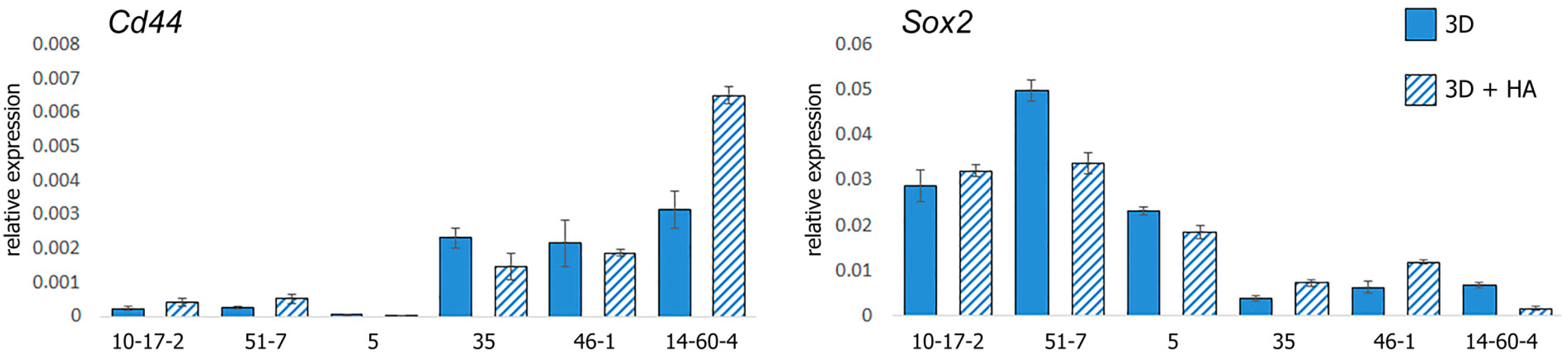
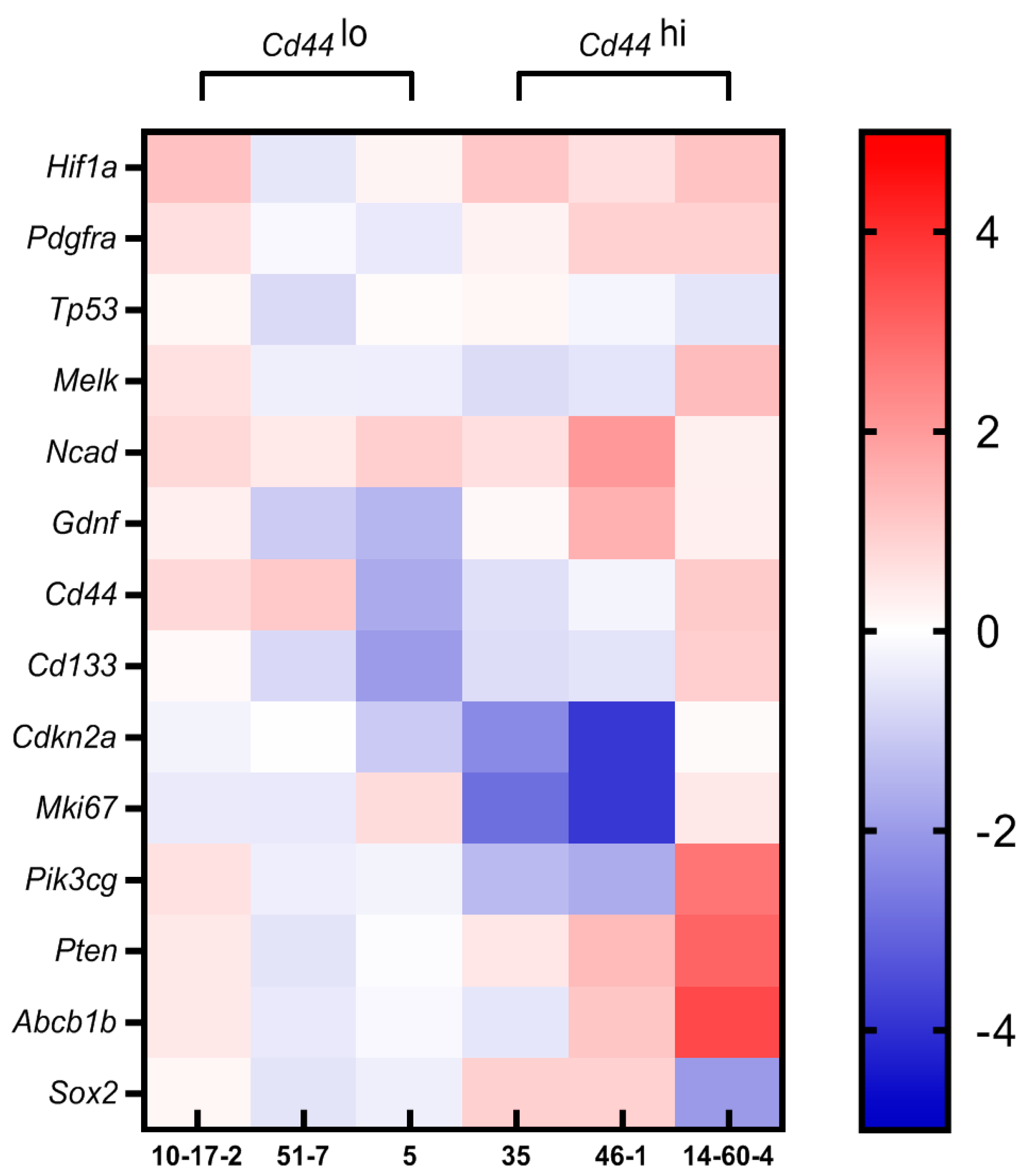
3.4. Gene Expression Profiling
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro-Oncology 2012, 14, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Barth, R.F.; Kaur, B. Rat brain tumor models in experimental neuro-oncology: The C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J. Neuro-Oncol. 2009, 94, 299–312. [Google Scholar] [CrossRef]
- Haddad, A.F.; Young, J.S.; Amara, D.; Berger, M.S.; Raleigh, D.R.; Aghi, M.K.; Butowski, N.A. Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neuro-Oncol. Adv. 2021, 3, vdab100. [Google Scholar] [CrossRef] [PubMed]
- Paolillo, M.; Comincini, S.; Schinelli, S. In vitro glioblastoma models: A journey into the third dimension. Cancers 2021, 13, 2449. [Google Scholar] [CrossRef]
- Hinderer, S.; Layland, S.L.; Schenke-Layland, K. ECM and ECM-like materials—Biomaterials for applications in regenerative medicine and cancer therapy. Adv. Drug Deliv. Rev. 2016, 97, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Song, I.; Dityatev, A. Crosstalk between glia, extracellular matrix and neurons. Brain Res. Bull. 2017, 136, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Abatangelo, G.; Vindigni, V.; Avruscio, G.; Pandis, L.; Brun, P. Hyaluronic Acid: Redefining Its Role. Cells 2020, 9, 1743. [Google Scholar] [CrossRef]
- Shahi, M.; Mohammadnejad, D.; Karimipour, M.; Rasta, S.H.; Rahbarghazi, R.; Abedelahi, A. Hyaluronic Acid and Regenerative Medicine: New Insights into the Stroke Therapy. Curr. Mol. Med. 2021, 20, 675–691. [Google Scholar] [CrossRef]
- Jensen, G.; Holloway, J.L.; Stabenfeldt, S.E. Hyaluronic Acid Biomaterials for Central Nervous System Regenerative Medicine. Cells 2020, 9, 2113. [Google Scholar] [CrossRef]
- Xiao, W.; Ehsanipour, A.; Sohrabi, A.; Seidlits, S.K. Hyaluronic-acid based hydrogels for 3-dimensional culture of patient-derived glioblastoma cells. J. Vis. Exp. 2018, 138, e58176. [Google Scholar] [CrossRef]
- Cannavà, C.; De Gaetano, F.; Stancanelli, R.; Venuti, V.; Paladini, G.; Caridi, F.; Ghica, C.; Crupi, V.; Majolino, D.; Ferlazzo, G.; et al. Chitosan-Hyaluronan Nanoparticles for Vinblastine Sulfate Delivery: Characterization and Internalization Studies on K-562 Cells. Pharmaceutics 2022, 14, 942. [Google Scholar] [CrossRef] [PubMed]
- Pedron, S.; Wolter, G.L.; Chen, J.-W.E.; Laken, S.E.; Sarkaria, J.N.; Harley, B.A. Hyaluronic acid-functionalized gelatin hydrogels reveal extracellular matrix signals temper the efficacy of erlotinib against patient-derived glioblastoma specimens. Biomaterials 2019, 219, 119371. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zhang, R.; Sohrabi, A.; Ehsanipour, A.; Sun, S.; Liang, J.; Walthers, C.M.; Ta, L.; Nathanson, D.A.; Seidlits, S.K. Brain-mimetic 3D culture platforms allow investigation of cooperative effects of extracellular matrix features on therapeutic resistance in glioblastoma. Cancer Res. 2018, 78, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Safarians, G.; Sohrabi, A.; Solomon, I.; Xiao, W.; Bastola, S.; Rajput, B.W.; Epperson, M.; Rosenzweig, I.; Tamura, K.; Singer, B.; et al. Glioblastoma Spheroid Invasion through Soft, Brain-Like Matrices Depends on Hyaluronic Acid–CD44 Interactions. Adv. Healthc. Mater. 2023, 12, e2203143. [Google Scholar] [CrossRef] [PubMed]
- Pibuel, M.A.; Poodts, D.; Díaz, M.; Hajos, S.E.; Lompardía, S.L. The scrambled story between hyaluronan and glioblastoma. J. Biol. Chem. 2021, 296, 100549. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Monni, E.; Congiu, T.; Massa, D.; Nat, R.; Diana, A. Human neurospheres: From stained sections to three-dimensional assembly. Transl. Neurosci. 2011, 2, 43–48. [Google Scholar] [CrossRef]
- Banerjee, D.; Singh, Y.P.; Datta, P.; Ozbolat, V.; O’Donnell, A.; Yeo, M.; Ozbolat, I.T. Strategies for 3D bioprinting of spheroids: A comprehensive review. Biomaterials 2022, 291, 121881. [Google Scholar] [CrossRef]
- Horder, H.; Lasheras, M.G.; Grummel, N.; Nadernezhad, A.; Herbig, J.; Ergün, S.; Teßmar, J.; Groll, J.; Fabry, B.; Bauer-Kreisel, P.; et al. Bioprinting and differentiation of adipose-derived stromal cell spheroids for a 3D breast cancer-adipose tissue model. Cells 2021, 10, 803. [Google Scholar] [CrossRef]
- Daly, A.C.; Davidson, M.D.; Burdick, J.A. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 2021, 12, 753. [Google Scholar] [CrossRef]
- Ding, Y.-W.; Zhang, X.-W.; Mi, C.-H.; Qi, X.-Y.; Zhou, J.; Wei, D.-X. Recent advances in hyaluronic acid-based hydrogels for 3D bioprinting in tissue engineering applications. Smart Mater. Med. 2023, 4, 59–68. [Google Scholar] [CrossRef]
- Nascimento, L.D.; Nicoletti, N.F.; Peletti-Figueiró, M.; Marinowic, D.; Falavigna, A. Hyaluronic Acid In Vitro Response: Viability and Proliferation Profile of Human Chondrocytes in 3D-Based Culture. Cartilage 2021, 13, 1077S–1087S. [Google Scholar] [CrossRef] [PubMed]
- Quílez, C.; Cerdeira, E.; González-Rico, J.; de Aranda, G.; López-Donaire, M.L.; Jorcano, J.L.; Velasco, D. Evaluation of different methodologies for primary human dermal fibroblast spheroid formation: Automation through 3D bioprinting technology. Biomed. Mater. 2022, 17, 055002. [Google Scholar] [CrossRef] [PubMed]
- Di Caprio, N.; Burdick, J.A. Engineered biomaterials to guide spheroid formation, function, and fabrication into 3D tissue constructs. Acta Biomater. 2023, 165, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Parra-Cantu, C.; Li, W.; Quiñones-Hinojosa, A.; Zhang, Y.S. 3D bioprinting of glioblastoma models. J. 3D Print. Med. 2020, 4, 113–125. [Google Scholar] [CrossRef]
- Navis, A.C.; Niclou, S.P.; Fack, F.; Stieber, D.; van Lith, S.; Verrijp, K.; Wright, A.; Stauber, J.; Tops, B.; Otte-Holler, I.; et al. Increased mitochondrial activity in a novel IDH1-R132H mutant human oligodendroglioma xenograft model: In situ detection of 2-HG and α-KG. Acta Neuropathol. Commun. 2013, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.C.; Koenderink, G.H.; Boukany, P.E. Spheroid mechanics and implications for cell invasion. Adv. Phys. X 2021, 6, 1978316. [Google Scholar] [CrossRef]
- Malgulwar, P.B.; Nambirajan, A.; Pathak, P.; Rajeshwari, M.; Suri, V.; Sarkar, C.; Singh, M.; Sharma, M.C. Epithelial-to-mesenchymal transition-related transcription factors are up-regulated in ependymomas and correlate with a poor prognosis. Hum. Pathol. 2018, 82, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.-G.; Oh, S.-J.; Ahn, E.-J.; Kim, Y.-J.; Jung, T.-Y.; Jung, S.; Kim, K.-K.; Lee, J.-H.; Lee, K.-H.; Moon, K.-S. Prognostic significance of E-cadherin and N-cadherin expression in Gliomas. BMC Cancer 2017, 17, 583. [Google Scholar] [CrossRef]
- Maherally, Z.; Smith, J.R.; Ghoneim, M.K.; Dickson, L.; An, Q.; Fillmore, H.L.; Pilkington, G.J. Silencing of CD44 in Glioma Leads to Changes in Cytoskeletal Protein Expression and Cellular Biomechanical Deformation Properties as Measured by AFM Nanoindentation. BioNanoScience 2015, 6, 54–64. [Google Scholar] [CrossRef]
- Kolliopoulos, C.; Ali, M.M.; Castillejo-Lopez, C.; Heldin, C.-H.; Heldin, P. CD44 Depletion in Glioblastoma Cells Suppresses Growth and Stemness and Induces Senescence. Cancers 2022, 14, 3747. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Wang, Y.; Liang, J.; Gao, S.; Cai, X.; Yu, Y.; Qi, Z.; Li, J.; Xie, Y.; Wang, Z. Association of glioma CD44 expression with glial dynamics in the tumour microenvironment and patient prognosis. Comput. Struct. Biotechnol. J. 2022, 20, 5203–5217. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-H.; Liao, C.-C.; Chow, N.-H.; Huang, L.L.-H.; Chuang, J.-I.; Wei, K.-C.; Shin, J.-W. Whether CD44 is an applicable marker for glioma stem cells. Am. J. Transl. Res. 2017, 9, 4785–4806. [Google Scholar] [PubMed]
- Ishii, H.; Mimura, Y.; Zahra, M.H.; Katayama, S.; Hassan, G.; Afify, S.M.; Seno, M. Isolation and characterization of cancer stem cells derived from human glioblastoma. Am. J. Cancer Res. 2021, 11, 441–457. [Google Scholar] [PubMed]
- Peng, L.; Fu, J.; Wang, W.; Hofman, F.M.; Chen, T.C.; Chen, L. Distribution of cancer stem cells in two human brain gliomas. Oncol. Lett. 2018, 17, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Abugomaa, A.; Elbadawy, M.; Yamanaka, M.; Goto, Y.; Hayashi, K.; Mori, T.; Uchide, T.; Azakami, D.; Fukushima, R.; Yoshida, T.; et al. Establishment of 2.5D organoid culture model using 3D bladder cancer organoid culture. Sci. Rep. 2020, 10, 9393. [Google Scholar] [CrossRef] [PubMed]
- Lemke, D.; Weiler, M.; Blaes, J.; Wiestler, B.; Jestaedt, L.; Klein, A.; Löw, S.; Eisele, G.; Radlwimmer, B.; Capper, D.; et al. Primary glioblastoma cultures: Can profiling of stem cell markers predict radiotherapy sensitivity? J. Neurochem. 2014, 131, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Tavianatou, A.G.; Caon, I.; Franchi, M.; Piperigkou, Z.; Galesso, D.; Karamanos, N.K. Hyaluronan: Molecular size-dependent signaling and biological functions in inflammation and cancer. FEBS J. 2019, 286, 2883–2908. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.-B.; Wang, N.-X.; Xu, Y.; Yu, C.-Y.; Liu, R.-M.; Luo, Y.; Xiao, J.-H. Hyaluronic acid ameliorates the proliferative ability of human amniotic epithelial cells through activation of TGF-β/BMP signaling. PeerJ 2020, 8, e10104. [Google Scholar] [CrossRef]
- Georgescu, M.-M. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer 2010, 1, 1170–1177. [Google Scholar] [CrossRef]
- Mukherjee, R.; Vanaja, K.G.; Boyer, J.A.; Gadal, S.; Solomon, H.; Chandarlapaty, S.; Levchenko, A.; Rosen, N. Regulation of PTEN translation by PI3K signaling maintains pathway homeostasis. Mol. Cell 2021, 81, 708–723.e5. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Ghatak, S.; Toole, B.P. Regulation of MDR1 Expression and drug resistance by a positive feedback loop involving hyaluronan, phosphoinositide 3-kinase, and ErbB2. J. Biol. Chem. 2005, 280, 20310–20315. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, L.Y.W.; Peyrollier, K.; Xia, W.; Gilad, E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J. Biol. Chem. 2008, 283, 17635–17651. [Google Scholar] [CrossRef] [PubMed]
- Gerardo-Ramírez, M.; Keggenhoff, F.L.; Giam, V.; Becker, D.; Groth, M.; Hartmann, N.; Straub, B.K.; Morrison, H.; Galle, P.R.; Marquardt, J.U.; et al. CD44 Contributes to the Regulation of MDR1 Protein and Doxorubicin Chemoresistance in Osteosarcoma. Int. J. Mol. Sci. 2022, 23, 8616. [Google Scholar] [CrossRef] [PubMed]
- Florczyk, S.J.; Wang, K.; Jana, S.; Wood, D.L.; Sytsma, S.K.; Sham, J.G.; Kievit, F.M.; Zhang, M. Porous chitosan-hyaluronic acid scaffolds as a mimic of glioblastoma microenvironment ECM. Biomaterials 2013, 34, 10143–10150. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.A.; Hartz, A.M.; Bauer, B. ABCB1 and ABCG2 Regulation at the Blood-Brain Barrier: Potential New Targets to Improve Brain Drug Delivery. Pharmacol. Rev. 2023, 75, 815–853. [Google Scholar] [CrossRef] [PubMed]
- Michalczyk, M.; Humeniuk, E.; Adamczuk, G.; Korga-Plewko, A. Hyaluronic Acid as a Modern Approach in Anticancer Therapy-Review. Int. J. Mol. Sci. 2022, 24, 103. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Moon, M.J.; Kim, D.Y.; Heo, S.H.; Jeong, Y.Y. Hyaluronic acid-based nanomaterials for cancer therapy. Polymers 2018, 10, 1133. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Wang, Y.; Ge, P.; Bailey, C.; Zhang, P.; Zhang, D.; Meng, Z.; Qi, C.; Chen, Q.; Chen, J.; et al. The HIF1α-PDGFD-PDGFRα axis controls glioblastoma growth at normoxia/mild-hypoxia and confers sensitivity to targeted therapy by echinomycin. J. Exp. Clin. Cancer Res. 2021, 40, 278. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, Y.; Xiong, S.; Williams-Villalobo, A.E. A Glance of p53 functions in brain development, neural stem cells, and brain cancer. Biology 2020, 9, 285. [Google Scholar] [CrossRef]
- Alekseeva, A.I.; Gerasimov, A.D.; Kudelkina, V.V.; Osipova, N.S.; Drozd, S.F.; Pavlova, G.V.; Kosyreva, A.M.; Fatkhudinov, T.K. Changes in Oncogene Expression in Experimental Glioblastoma 101.8 Rats during Therapy with PLGA Nanoparticles Loaded with Doxorubicin. Bull. Exp. Biol. Med. 2023, 174, 518–522. [Google Scholar] [CrossRef] [PubMed]




| Tissue Strain | Tumor Type | IHC Markers | Animal Species | Carcinogen |
|---|---|---|---|---|
| 10-17-2 | Astrocytoma | GFAP+ ATRX− CD68− | Rattus norvegicus, rat | N-ethyl-N-nitrosourea |
| 51-7 | oligodendroglioma | ATRX+ | Mus musculus, mouse | 7,12-Dimethylbenzathracene |
| 5 | Ependymoma | GFAP+ CD99+ | Mus musculus, mouse | 7,12-Dimethylbenzathracene |
| 35 | Glioblastoma | Iba1+ CD68+ Cathepsin B+ | Rattus norvegicus, rat | N-Nitrosomethylurea |
| 46-1 | Neuroma | Calretinin+ CD56+ S-100+ | Rattus norvegicus, rat | N-ethyl-N-nitrosourea |
| 14-60-4 | Glioblastoma | Iba1+ CD68+ | Rattus norvegicus, rat | N-ethyl-N-nitrosourea |
| Gene | Rat | Mouse |
|---|---|---|
| Hif1a | (5′ to 3′) TCACAGTCGGACAACCTCAC (3′ to 5′) TGCTGCAGTAACGTTCCAATTC | (5′ to 3′) AGGATGAGTTCTGAACGTCGAAA (3′ to 5′) CTGTCTAGACCACCGGCATC |
| Pdgfra | (5′ to 3′) ATTGTGCTCCGTAGTCCCCA (3′ to 5′) TCCTAGCCAGGGATCTTGCC | (5′ to 3′) GTGCTAGCGCGGAACCT (3′ to 5′) CATAGCTCCTGAGACCCGC |
| Tp53 | (5′ to 3′) AGGGAGTGCAAAGAGAGCAC (3′ to 5′) GTCTTCGGGTAGCTGGAG G | (5′ to 3′) TTCTCCGAAGACTGGATGACTG (3′ to 5′) TCCATGCAGTGAGGTGATGG |
| Melk | (5′ to 3′) TGCCTTGGGCAGAAAAACAGAT (3′ to 5′) GGCCTGCAGGTCGGATACT | (5′ to 3′) ACCAAGAAAGCGAAAGCTGC (3′ to 5′) CGAAGTCCACGTTCTTCTTTGG |
| Ncad | (5′ to 3′) ACCCAGGAAAAGTGGCAGGT (3′ to 5′) TCGGAGGGATGACCCAGTCT | (5′ to 3′) TGTGGAGGCTTCTGGTGAAAT (3′ to 5′) TCTCACAGCATACACCGTGC |
| Ecad | (5′ to 3′) CCTTCCTCCCAATACATCTCCC (3′ to 5′) TCTCCGCCTCCTTCTTCATC | (5′ to 3′) GGTTTTCTACAGCATCACCG (3′ to 5′) GCTTCCCCATTTGATGACAC |
| Gdnf | (5′ to 3′) TATGGGATGTCGTGGCTGTCT (3′ to 5′) AGTCACTGGTCAGCGCGAAG | (5′ to 3′) GACCGGATCCGAGGTGC (3′ to 5′) GAGGGAGTGGTCTTCAGCG |
| Cd44 | (5′ to 3′) TTTATTGGGAGCACCCTGGC (3′ to 5′) AGGGGTAGTCATCAAGGCTGT | (5′ to 3′) AGAAGGGACAACTGCTTCGG (3′ to 5′) TTGGAGCTGCAGTAGGCTG |
| Cd133 | (5′ to 3′) AATTCAAAGCCCCGGGAGTG (3′ to 5′) CTGGGGGATCAGGCCAACTA | (5′ to 3′) GGAGCAGTACACCAACACCA (3′ to 5′) GTCTGTTTGATGGCTGTCGC |
| Cdkn2a | (5′ to 3′) GTACCCCGATACAGGTGATGAT (3′ to 5′) GATACCGCAAATACCGCACG | (5′ to 3′) TGGTCACTGTGAGGATTCAGC (3′ to 5′) TGCCCATCATCATCACCTGG |
| Mki67 | (5′ to 3′) TTCCAGACACCAGACCATGC (3′ to 5′) GGGTTCTAACTGGTCTTC TGG | (5′ to 3′) CCTGCCTGTTTGGAAGGAGT (3′ to 5′) AAGGAGCGGTCAATGATGGTT |
| Pik3cg | (5′ to 3′) TGGATATGAAGGGAGCCCCA (3′ to 5′) CATGCCCTAGGTGACCTGAC | (5′ to 3′) CCGCTCAGGGAGAGGAGTA (3′ to 5′) CCACTCTCAGCTTCACCTCC |
| Pten | (5′ to 3′) GCCAAATTTAACTGCAGAGTTGC (3′ to 5′) GCGCCTCTGACTGGGAATAG | (5′ to 3′) GGACCAGAGACAAAAAGGGAGT (3′ to 5′) CCTTTAGCTGGCAGACCACA |
| Abcb1b | (5′ to 3′) GTCCACCATCCA AACGCAG (3′ to 5′) GCACCTCAAATACTCCCAGCT C | (5′ to 3′) CTCTTGAAGCCGTAAGAGGCT (3′ to 5′) AACTCCATCACCACCTCACG |
| Sox2 | (5′ to 3′) TCCATGGGCTCTGTG GTCAA (3′ to 5′) CATGTGCAGTCTACTGGGCG | (5′ to 3′) AGGAAAGGGTTCTTGCTGGG (3′ to 5′) GGTCTTGCCAGTACTTGCTCT |
| Gapdh | (5′ to 3′) CAGGGCTGCCTTCTCTTGTG (3′ to 5′) GCCTTGACTGTGCCGTTGAA | (5′ to 3′) AACTCAGGAGAGTGTTTCCTCG (3′ to 5′) TTTGCCGTGAGTGGAGTCAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arutyunyan, I.; Soboleva, A.; Balchir, D.; Jumaniyazova, E.; Kudelkina, V.; Elchaninov, A.; Fatkhudinov, T. Hyaluronic Acid Prevents Fusion of Brain Tumor-Derived Spheroids and Selectively Alters Their Gene Expression Profile. Biomolecules 2024, 14, 466. https://doi.org/10.3390/biom14040466
Arutyunyan I, Soboleva A, Balchir D, Jumaniyazova E, Kudelkina V, Elchaninov A, Fatkhudinov T. Hyaluronic Acid Prevents Fusion of Brain Tumor-Derived Spheroids and Selectively Alters Their Gene Expression Profile. Biomolecules. 2024; 14(4):466. https://doi.org/10.3390/biom14040466
Chicago/Turabian StyleArutyunyan, Irina, Anna Soboleva, Dorzhu Balchir, Enar Jumaniyazova, Vera Kudelkina, Andrey Elchaninov, and Timur Fatkhudinov. 2024. "Hyaluronic Acid Prevents Fusion of Brain Tumor-Derived Spheroids and Selectively Alters Their Gene Expression Profile" Biomolecules 14, no. 4: 466. https://doi.org/10.3390/biom14040466






