Enhancing Selenium Accumulation in Rhodotorula mucilaginosa Strain 6S Using a Proteomic Approach for Aquafeed Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Soil Samples and Isolation of Yeast
2.2. DNA Extraction and Amplification of the ITS rDNA Region
2.3. Molecular Identification and Phylogenetic Analysis
2.4. Preparation of the Inoculum and Strain Maintenance
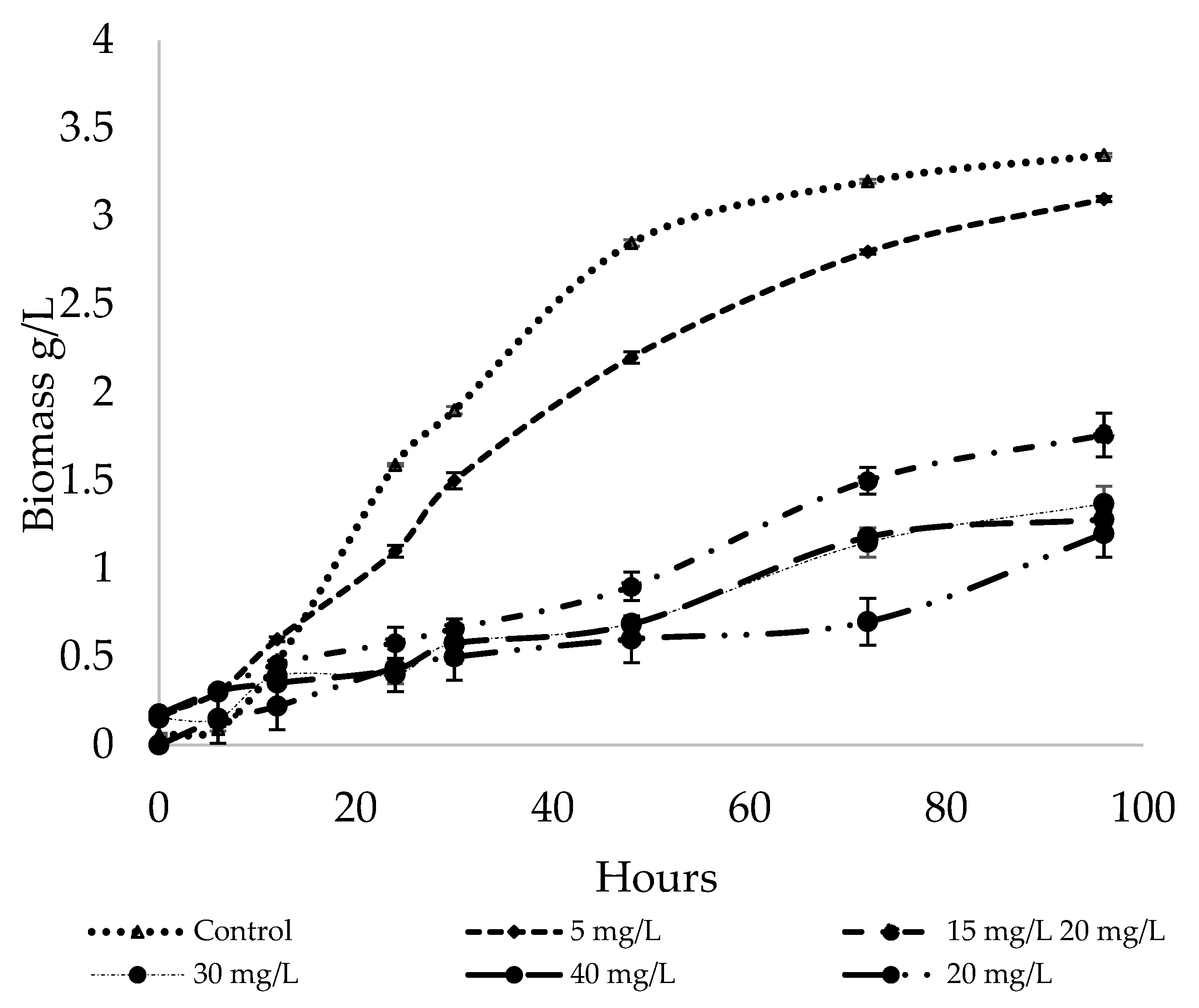
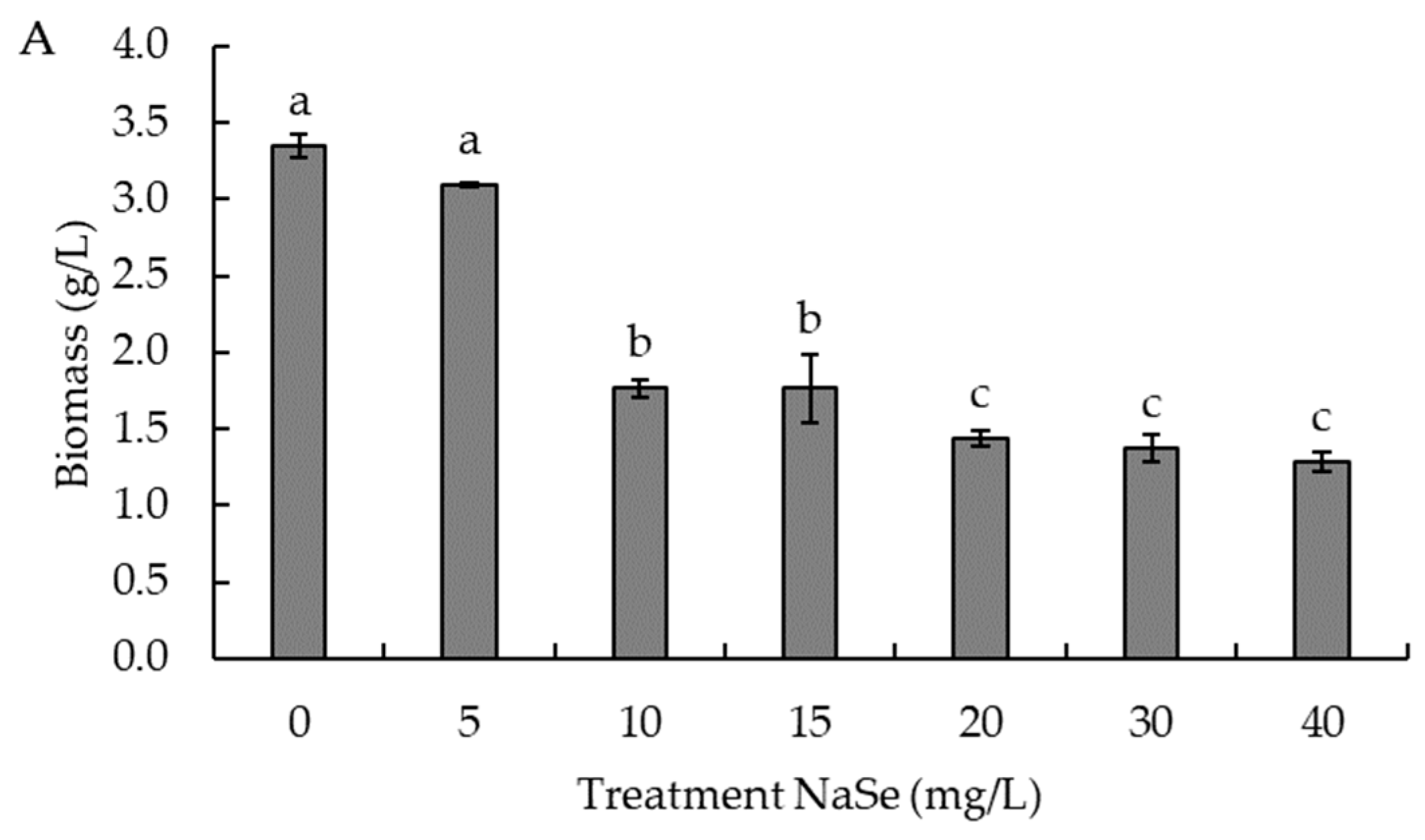
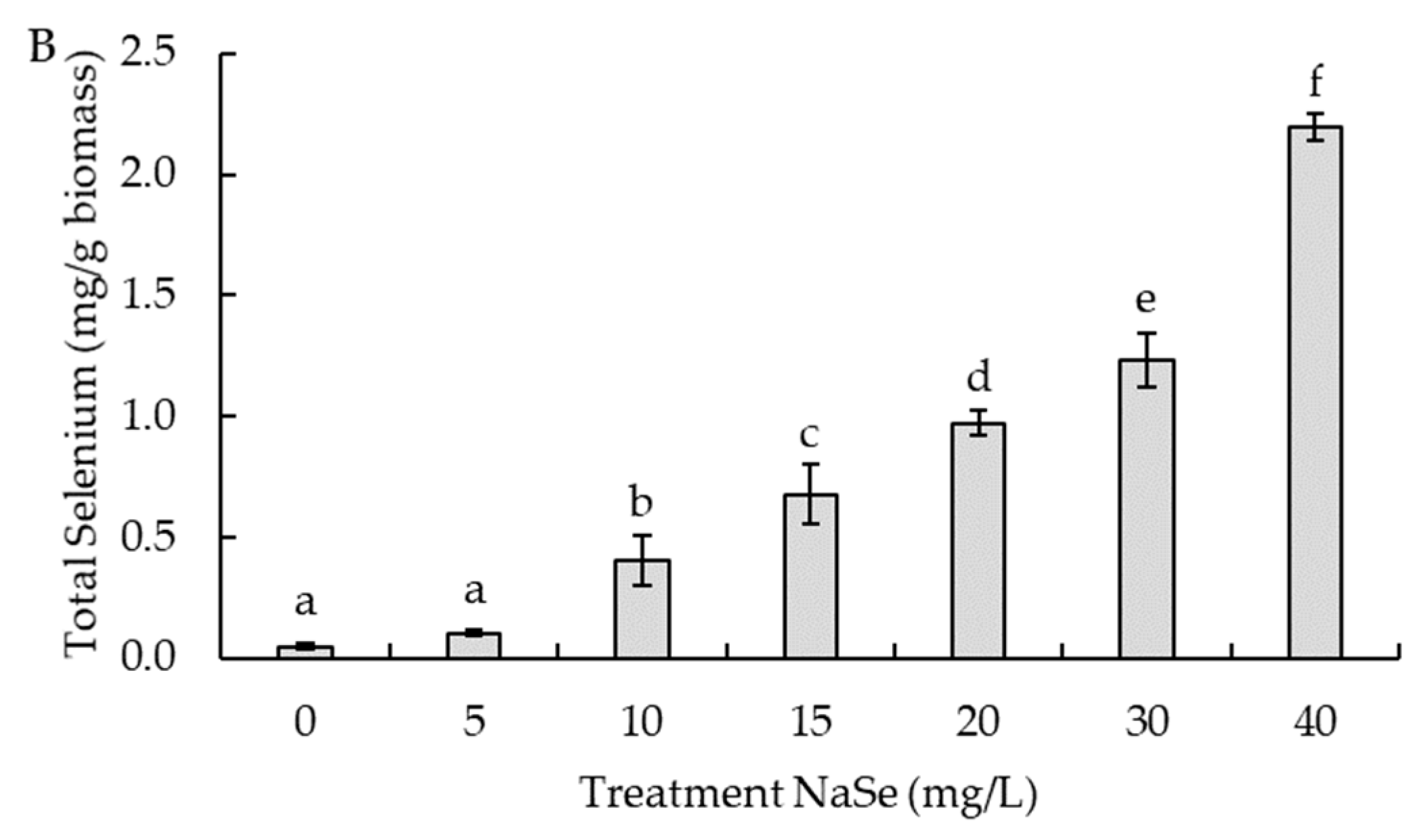
2.5. Effect of Sodium Selenite Concentration on Selenium Accumulation and Cell Growth
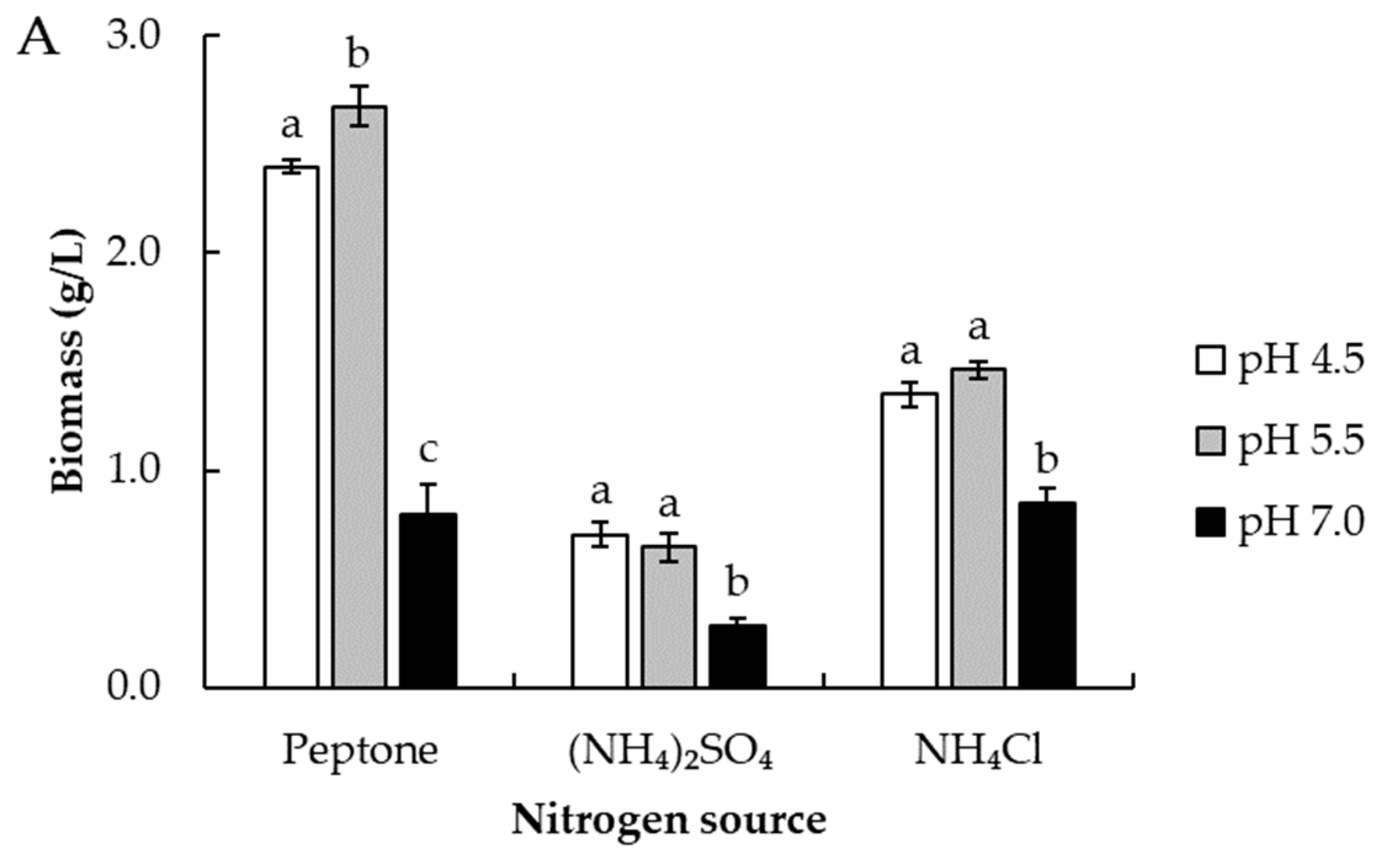
2.6. Effect of pH and Nitrogen Source on Selenium Accumulation and Cell Growth
2.7. Optimization in a 2 L Bioreactor and Nutritional Characterization of Biomass
2.8. Protein Extraction and Analysis of Extracellular Fractions
Proteome Data Processing
2.9. Analytical Methods
2.10. Statistical Analysis
3. Results
3.1. Identification of the Selected Yeast
3.2. Effect of Sodium Selenite Concentration
3.3. Effect of pH and Nitrogen Source on Growth and Total Selenium Accumulation
3.4. Cultivation in a 2 L Reactor and Biomass Characterization
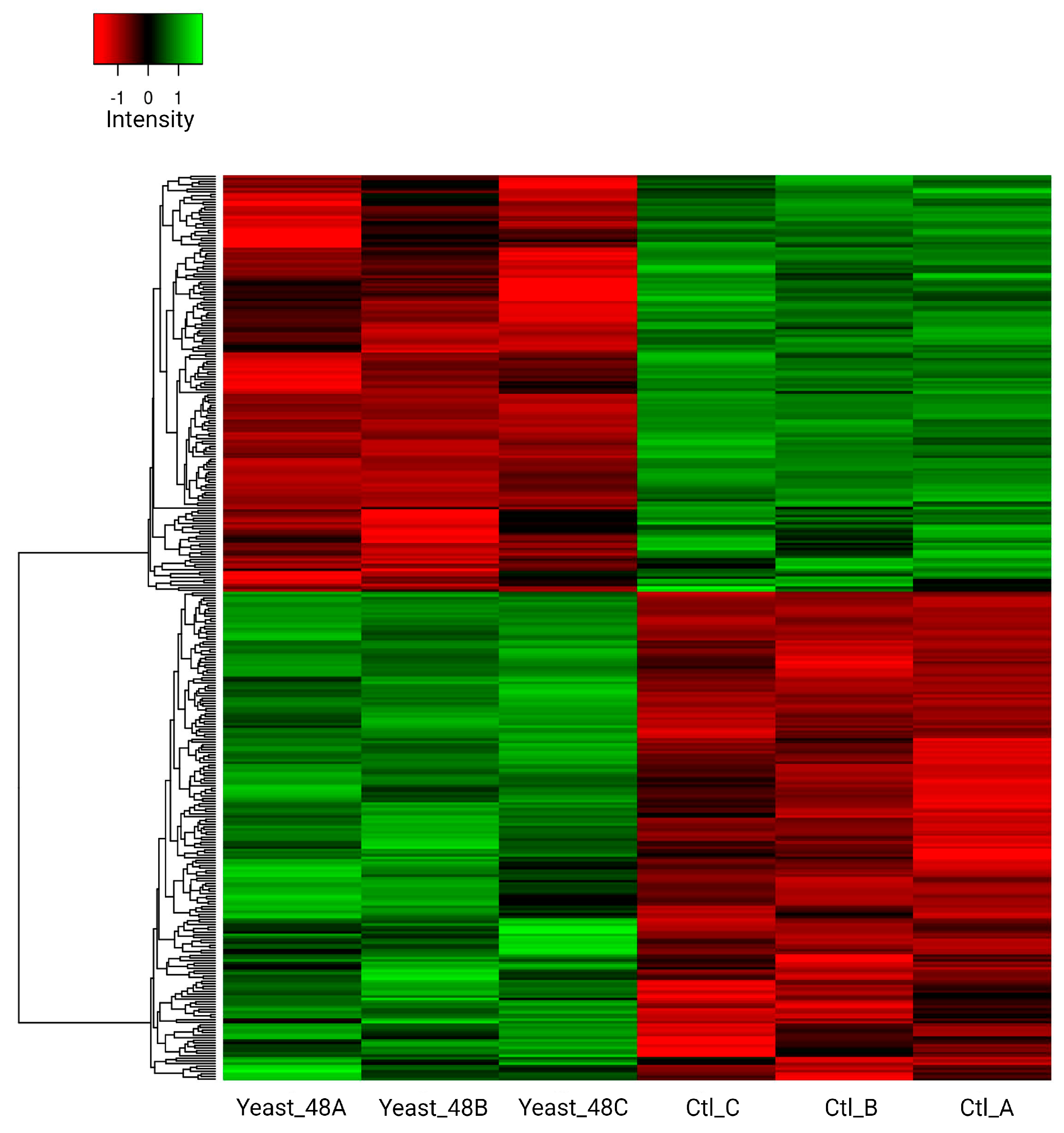
3.5. Proteome R. mucilaginosa 6S
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farzad, R.; Kuhn, D.D.; Smith, S.A.; O’Keefe, S.F.; Hines, I.S.; Bushman, T.J.; Galagarza, O.A.; Stevens, A.M. Effects of Selenium-Enriched Prebiotic on the Growth Performance, Innate Immune Response, Oxidative Enzyme Activity and Microbiome of Rainbow Trout (Oncorhynchus mykiss). Aquaculture 2021, 531, 735980. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błazejak, S. Selenium: Significance, and Outlook for Supplementation. Nutrition 2013, 29, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Alehagen, U.; Opstad, T.B.; Alexander, J.; Larsson, A.; Aaseth, J. Impact of Selenium on Biomarkers and Clinical Aspects Related to Ageing. A Review. Biomolecules 2021, 11, 1478. [Google Scholar] [CrossRef] [PubMed]
- González-Salitre, L.; Román-Gutiérrez, A.; Contreras-López, E.; Bautista-Ávila, M.; Rodríguez-Serrano, G.; González-Olivares, L. Promising Use of Selenized Yeast to Develop New Enriched Food: Human Health Implications. Food Rev. Int. 2023, 39, 1594–1611. [Google Scholar] [CrossRef]
- Long, M.; Lin, W.; Hou, J.; Guo, H.; Li, L.; Li, D. Dietary Supplementation with Selenium Yeast and Tea Polyphenols Improve Growth Performance and Nitrite Tolerance of Wuchang Bream (Megalobrama amblycephala). Fish Shellfish. Immunol. 2017, 68, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Sumana, S.L.; Chen, H.; Shui, Y.; Zhang, C.; Yu, F.; Zhu, J.; Su, S. Effect of Dietary Selenium on the Growth and Immune Systems of Fish. Animals 2023, 13, 2978. [Google Scholar] [CrossRef] [PubMed]
- Pacitti, D.; Lawan, M.M.; Sweetman, J.; Martin, S.A.M.; Feldmann, J.; Secombes, C.J. Selenium Supplementation in Fish: A Combined Chemical and Biomolecular Study to Understand Sel-Plex Assimilation and Impact on Selenoproteome Expression in Rainbow Trout (Oncorhynchus mykiss). PLoS ONE 2015, 10, e0127041. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.L.; Haslam, R.P.; Napier, J.A.; Sayanova, O. Metabolic Engineering of Phaeodactylum Tricornutum for the Enhanced Accumulation of Omega-3 Long Chain Polyunsaturated Fatty Acids. Metab. Eng. 2014, 22, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Oraby, M.M.; Allababidy, T.; Ramadan, E.M. The Bioavailability of Selenium in Saccharomyces Cerevisiae. Ann. Agric. Sci. 2015, 60, 307–315. [Google Scholar] [CrossRef]
- Díaz-Navarrete, P.; Dantagnan, P.; Henriquez, D.; Soto, R.; Correa-Galeote, D.; Sáez-Arteaga, A. Selenized Non-Saccharomyces Yeasts and Their Potential Use in Fish Feed. Fish Physiol. Biochem. 2024; Online ahead of print. [Google Scholar] [CrossRef]
- Kieliszek, M. Spectrophotometric Evaluation of Selenium Binding by Saccharomyces Cerevisiae ATCC MYA-2200 and Candida Utilis ATCC 9950 Yeast. J. Trace Elem. Med. Biol. 2016, 35, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Hozien, S.T.; Abdel Wahab, M.M.; Hassan, A.M. Ameliorative Effect of Selenium Yeast Supplementation on the Physio-Pathological Impacts Ofchronic Exposure to Glyphosate and or Malathion in Oreochromis Niloticus. BMC Vet. Res. 2022, 18, 159. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hu, J.; Zhang, Y.; Bai, L. Evaluation of Largemouth Bass (Micropterus salmoide) Fed Selenium Yeast Diets: Growth, Histopathology, Antioxidant Ability, and Apoptosis. Aquac. Rep. 2023, 29, 101505. [Google Scholar] [CrossRef]
- Otero, C.; ICIDCA. Sobre Los Derivados de La Caña de Azúcar Enriquecimiento de Biomasa de Levadura Con Micronutrientes Esenciales ICIDCA. Sobre Los Deriv. Caña 2008, XLII, 60–68. [Google Scholar]
- Bhattacharjee, A.; Basu, A.; Bhattacharya, S. Selenium Nanoparticles Are Less Toxic than Inorganic and Organic Selenium to Mice in Vivo. Nucleus 2019, 62, 259–268. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, K.S. Role of Nano-Selenium in Health and Environment. J. Biotechnol. 2021, 325, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Waśko, A.; Michalak, K.; Kot, A.M.; Piwowarek, K.; Winiarczyk, S. Effect of Selenium and Methods of Protein Extraction on the Proteomic Profile of Saccharomyces Yeast. Open Life Sci. 2022, 17, 1117–1128. [Google Scholar] [CrossRef]
- Ponce de León, C.A.; Bayón, M.M.; Paquin, C.; Caruso, J.A. Selenium Incorporation into Saccharomyces cerevisiae Cells: A Study of Different Incorporation Methods. J. Appl. Microbiol. 2002, 92, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Q.; Zhao, W.; Xie, S.W.; Xie, J.J.; Zhang, Z.H.; Tian, L.X.; Liu, Y.J.; Niu, J. Effects of Dietary Hydrolyzed Yeast (Rhodotorula mucilaginosa) on Growth Performance, Immune Response, Antioxidant Capacity and Histomorphology of Juvenile Nile Tilapia (Oreochromis niloticus). Fish Shellfish. Immunol. 2019, 90, 30–39. [Google Scholar] [CrossRef]
- Pankiewicz, U.; Jamroz, J.; Schodziñski, A. Optimization of Selenium Accumulation in Rhodotorula Rubra Cells by Treatment of Culturing Medium with Pulse Electric Field. Int. Agrophys. 2006, 4, 147–152. [Google Scholar]
- Ruocco, M.H.W.; Chan, C.S.; Hanson, T.E.; Church, T.M. Characterization and Distribution of Selenite Reduction Products in Cultures of the Marine Yeast Rhodotorula mucilaginosa-13B. Geomicrobiol. J. 2014, 31, 769–778. [Google Scholar] [CrossRef]
- González-Salitre, L.; Castañeda-Ovando, A.; Basilio-Cortés, U.A.; del Carmen García-Contreras, A.; Rodríguez Serrano, G.M.; Cardelle-Cobas, A.; Román-Gutiérrez, A.D.; González-Olivares, L.G. Biogenic Production of Seleno-Amino Acids and Seleno-Nanoparticles by Saccharomyces boulardii. Food Biosci. 2023, 53, 102552. [Google Scholar] [CrossRef]
- Kieliszek, M.; Bierla, K.; Jiménez-Lamana, J.; Kot, A.M.; Alcántara-Durán, J.; Piwowarek, K.; Błażejak, S.; Szpunar, J. Metabolic Response of the Yeast Candida utilis during Enrichment in Selenium. Int. J. Mol. Sci. 2020, 21, 5287. [Google Scholar] [CrossRef] [PubMed]
- Díaz, P.E.; Aranda, C.; Martínez, O.; Godoy, R.; Gonzales, A.; Valenzuela, E. Characterization of Yeast in Hapludands Soil with Biotechnological Potential. J. Soil Sci. Plant Nutr. 2017, 17, 948–965. [Google Scholar] [CrossRef]
- Díaz-Navarrete, P.; Marileo, L.; Madrid, H.; Belezaca-Pinargote, C.; Dantagnan, P. Lipid Production from Native Oleaginous Yeasts Isolated from Southern Chilean Soil Cultivated in Industrial Vinasse Residues. Microorganisms 2023, 11, 2516. [Google Scholar] [CrossRef] [PubMed]
- Altschup, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. (JMB) 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Scorzetti, G.; Fell, J.W.; Fonseca, A.; Statzell-Tallman, A. Systematics of Basidiomycetous Yeasts: A Comparison of Large Subunit D1/D2 and Internal Transcribed Spacer RDNA Regions. FEMS Yeast Res. 2002, 2, 495–517. [Google Scholar] [CrossRef] [PubMed]
- Civiero, E.; Pintus, M.; Ruggeri, C.; Tamburini, E.; Sollai, F.; Sanjust, E.; Zucca, P. Physiological and Phylogenetic Characterization of Rhodotorula Diobovata DSBCA06, a Nitrophilous Yeast. Biology 2018, 7, 39. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Piwowarek, K.; Brzezicka, K. Equilibrium Modeling of Selenium Binding from Aqueous Solutions by Candida Utilis ATCC 9950 Yeasts. 3 Biotech 2018, 8, 388. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Kot, A.M.; Kolotylo, V. Bioaccumulation of Selenium and Production of Carotenoids by the Yeast Rhodotorula mucilaginosa. Biocatal. Agric. Biotechnol. 2023, 53, 102903. [Google Scholar] [CrossRef]
- Janzs, B.; Pais, I.; Vereczkey, G. Trace Elements Preparation of Selenium Yeasts I. Preparation of Selenium-Enriched Saccharomyces Cerevisiae. J. Trace Elements Med. Biol. 2000, 14, 43–47. [Google Scholar]
- Folch, J.; Lees, M.; Sloane, G.H. A Simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Morrison, W.R.; Smith, L.M. Preparation of Fatty Acid Methyl Esters and Dimethylacetals from Lipids with Boron Fluoride-Methanol. J. Lipid Res. 1964, 5, 600–608. [Google Scholar] [CrossRef]
- Garcia-Cortes, A.; Garcia-Vásquez, J.A.; Aranguren, Y.; Ramirez-Castrillon, M. Pigment Production Improvement in Rhodotorula Mucilaginosa Ajb01 Using Design of Experiments. Microorganisms 2021, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, A.A.; Gadela, R.; Bharali, B.; Deshavath, N.N.; Dasu, V.V. Development of High Biomass and Lipid Yielding Medium for Newly Isolated Rhodotorula Mucilaginosa. Fuel 2019, 239, 874–885. [Google Scholar] [CrossRef]
- Aksu, Z.; Tuǧba Eren, A. Carotenoids Production by the Yeast Rhodotorula Mucilaginosa: Use of Agricultural Wastes as a Carbon Source. Process Biochem. 2005, 40, 2985–2991. [Google Scholar] [CrossRef]
- Li, Z.; Li, C.; Cheng, P.; Yu, G. Rhodotorula Mucilaginosa—Alternative Sources of Natural Carotenoids, Lipids, and Enzymes for Industrial Use. Heliyon 2022, 8, e11505. [Google Scholar] [CrossRef]
- Brunel, M.; Burkina, V.; Pickova, J.; Sampels, S.; Moazzami, A.A. Oleaginous Yeast Rhodotorula Toruloides Biomass Effect on the Metabolism of Arctic Char (Salvelinus alpinus). Front. Mol. Biosci. 2022, 9, 931946. [Google Scholar] [CrossRef]
- Marinescu, G.; Gabriela Stoicescu, A.; Teodorof, L. Industrial nutrient medium use for yeast selenium preparation. Food Technol. 2011, 35, 45–53. [Google Scholar]
- Schrauzer, G.N. Selenium Yeast: Composition, Quality, Analysis, and Safety. Pure Appl. Chem. 2006, 78, 105–109. [Google Scholar] [CrossRef]
- Sun, Y.; Gan, Y.; Zhang, L.; Shi, Y.; Yue, T.; Yuan, Y. Isolation and Identification of Monascus and Evaluation of Its Selenium Accumulation. LWT 2022, 154, 112887. [Google Scholar] [CrossRef]
- Pinson, B.; Sagot, I.; Daignan-Fornier, B. Identification of Genes Affecting Selenite Toxicity and Resistance in Saccharomyces Cerevisiae. Mol. Microbiol. 2000, 36, 679–687. [Google Scholar] [CrossRef]
- Princová, J.; Salat-Canela, C.; Daněk, P.; Marešová, A.; de Cubas, L.; Bähler, J.; Ayté, J.; Hidalgo, E.; Převorovský, M. Perturbed Fatty-Acid Metabolism Is Linked to Localized Chromatin Hyperacetylation, Increased Stress-Response Gene Expression and Resistance to Oxidative Stress. PLoS Genet. 2023, 19, 1010582. [Google Scholar] [CrossRef]
- Perrone, G.G.; Tan, S.X.; Dawes, I.W. Reactive Oxygen Species and Yeast Apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Martiniano, S.E.; Philippini, R.R.; Franco-Marcelino, P.R.; da Silva, S.S. Effect of Selenium Uptake on Growth Metabolism in Yeasts for the Production of Enriched Single-Cell Protein Using Agro-Industrial by-Products. Biomass. Convers. Biorefinery 2022, 12, 3975–3983. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Bzducha-Wróbel, A.; Kot, A.M. Effect of Selenium on Lipid and Amino Acid Metabolism in Yeast Cells. Biol. Trace Elem. Res. 2019, 187, 316–327. [Google Scholar] [CrossRef]
- Nie, X.; Zhu, Z.; Lu, H.; Xue, M.; Tan, Z.; Zhou, J.; Xin, Y.; Mao, Y.; Shi, H.; Zhang, D. Assembly of Selenium Nanoparticles by Protein Coronas Composed of Yeast Protease A. Process Biochem. 2023, 129, 140–149. [Google Scholar] [CrossRef]
- Miletić, D.; Turło, J.; Podsadni, P.; Sknepnek, A.; Szczepańska, A.; Klimaszewska, M.; Malinowska, E.; Lević, S.; Nedović, V.; Nikšić, M. Production of Bioactive Selenium Enriched Crude Exopolysaccharides via Selenourea and Sodium Selenite Bioconversion Using Trametes Versicolor. Food Biosci. 2021, 42, 101046. [Google Scholar] [CrossRef]
- Ogra, Y.; Shimizu, M.; Takahashi, K.; Anan, Y. Biotransformation of Organic Selenium Compounds in Budding Yeast, Saccharomyces cerevisiae. Metallomics 2018, 10, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, L.; Tan, T. High Cell Density Fermentation of Saccharomyces Cerevisiae GS2 for Selenium-Enriched Yeast Production. Korean J. Chem. Eng. 2010, 27, 1836–1840. [Google Scholar] [CrossRef]
- Kitajima, T.; Chiba, Y. Selenomethionine Metabolism and Its Toxicity in Yeast. Biomol. Concepts 2013, 4, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Čertík, M.; Breierová, E.; Oláhová, M.; Šajbidor, J.; Márová, I. Effect of Selenium on Lipid Alternations in Pigment-Forming Yeasts. Food Sci. Biotechnol. 2013, 22, 45–51. [Google Scholar] [CrossRef]
- Mapelli, V.; Hillestrøm, P.R.; Kápolna, E.; Larsen, E.H.; Olsson, L. Metabolic and Bioprocess Engineering for Production of Selenized Yeast with Increased Content of Seleno-Methylselenocysteine. Metab. Eng. 2011, 13, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, Z.; Wang, C.; Li, E.; Qin, J.G.; Chen, L. Fish and Shell Fi Sh Immunology Dietary Supplementation of Selenium Yeast Enhances the Antioxidant Capacity and Immune Response of Juvenile Eriocheir Sinensis under Nitrite Stress. Fish Shellfish. Immunol. 2019, 87, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Marinescu, G.; Stoicescu, A.G.; Teodorof, L. Industrial nutrient medium use for yeast selenium preparation. Ann. Univ. Dunarea De Jos Galati. Fascicle VI. Food Technol. 2011, 35, 45. [Google Scholar]
- Yao, G.; Wang, W.; Ao, L.; Cheng, Z.; Wu, C.; Pan, Z.; Liu, K.; Li, H.; Su, W.; Fang, L. Improved Total Synthesis and Biological Evaluation of Coibamide A Analogues. J. Med. Chem. 2018, 60, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Wellinger, R. Yeast as a Model System to Study Metabolic Impact of Selenium Compounds. Microb. Cell 2015, 2, 139–149. [Google Scholar] [CrossRef]
- Losi, M.E.; Frankenberger, W.T. Reduction of Selenium Oxyanions by Enterobacter Cloacae SLD1a-1: Isolation and Growth of the Bacterium and Its Expulsion of Selenium Particles. Appl. Environ. Microbiol. 1997, 63, 3079–3084. [Google Scholar] [CrossRef]
- Breierová, E.; Gregor, T.; Marova, I.; Čertík, M.; Kogan, G. Enhanced Antioxidant Formula Based on a Selenium-Supplemented Carotenoid-Producing Yeast Biomass. Chem. Biodivers. 2008, 5, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Polburee, P.; Yongmanitchai, W.; Lertwattanasakul, N.; Ohashi, T.; Fujiyama, K.; Limtong, S. Characterization of Oleaginous Yeasts Accumulating High Levels of Lipid When Cultivated in Glycerol and Their Potential for Lipid Production from Biodiesel-Derived Crude Glycerol. Fungal Biol. 2015, 119, 1194–1204. [Google Scholar] [CrossRef]
- Maldonade, I.R.; Rodriguez-Amaya, D.B.; Scamparini, A.R.P. Statistical optimisation of cell growth and carotenoid production by rhodotorula mucilaginosa. Braz. J. Microbiol. 2012, 43, 109–115. [Google Scholar] [CrossRef]
- Kieliszek, M.; Dourou, M. Effect of Selenium on the Growth and Lipid Accumulation of Yarrowia lipolytica Yeast. Biol. Trace Elem. Res. 2011, 199, 1611–1622. [Google Scholar] [CrossRef] [PubMed]
- Barchielli, G.; Capperucci, A.; Tanini, D. The Role of Selenium in Pathologies: An Updated Review. Antioxidants 2022, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Feron, G.; Blin-Perrin, C.; Krasniewski, I.; Mauvais, G.; Lherminier, J. Metabolism of Fatty Acid in Yeast: Characterisation of β-Oxidation and Ultrastructural Changes in the Genus Sporidiobolus sp. Cultivated on Ricinoleic Acid Methyl Ester. FEMS Microbiol. Lett. 2005, 250, 63–69. [Google Scholar] [CrossRef]
- Van Roermund, C.W.T.; Waterham, H.R.; Ijlst, L.; Wanders, R.J.A. Fatty Acid Metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 2003, 60, 1838–1851. [Google Scholar] [CrossRef]
- Mapelli, V.; Hillestrøm, P.R.; Patil, K.; Larsen, E.H.; Olsson, L. The Interplay between Sulphur and Selenium Metabolism Influences the Intracellular Redox Balance in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 20–32. [Google Scholar] [CrossRef]
- Ferrari, L.; Cattaneo, D.M.I.R.; Abbate, R.; Manoni, M.; Ottoboni, M.; Luciano, A.; von Holst, C.; Pinotti, L. Advances in Selenium Supplementation: From Selenium-Enriched Yeast to Potential Selenium-Enriched Insects, and Selenium Nanoparticles. Anim. Nutr. 2023, 14, 193–203. [Google Scholar] [CrossRef]
- Irazusta, V.; Estévez, C.; Amoroso, M.J.; De Figueroa, L.I.C. Proteomic Study of the Yeast Rhodotorula Mucilaginosa RCL-11 under Copper Stress. BioMetals 2012, 25, 517–527. [Google Scholar] [CrossRef]
- Ilyas, S.; Rehman, A.; Coelho, A.V.; Sheehan, D. Proteomic Analysis of an Environmental Isolate of Rhodotorula Mucilaginosa after Arsenic and Cadmium Challenge: Identification of a Protein Expression Signature for Heavy Metal Exposure. J. Proteom. 2016, 141, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Landolfo, S.; Abbondio, M.; Cutzu, R.; Biosa, G.; Pagnozzi, D.; Uzzau, S.; Mannazzu, I. Proteomic Analysis of Rhodotorula Mucilaginosa: Dealing with the Issues of a Non-Conventional Yeast. Yeast 2016, 33, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Błażejak, S.; Gientka, I.; Bzducha-Wróbel, A. Accumulation and Metabolism of Selenium by Yeast Cells. Appl. Microbiol. Biotechnol. 2015, 99, 5373–5382. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, N.; Zhou, Y.; Godana, E.A.; Dhanasekaran, S.; Gu, X.; Zhao, L.; Zhang, H. Transcriptome Analysis Reveals the Mechanisms Involved in the Enhanced Antagonistic Efficacy of Rhodotorula mucilaginosa Induced by Chitosan. LWT 2021, 142, 110992. [Google Scholar] [CrossRef]
- Castañeda-Tamez, P.; Chiquete-Félix, N.; Uribe-Carvajal, S.; Cabrera-Orefice, A. The Mitochondrial Respiratory Chain from Rhodotorula mucilaginosa, an Extremophile Yeast. Biochim. Biophys. Acta (BBA) Bioenerg. 2024, 1865, 149035. [Google Scholar] [CrossRef]
- Grigore, D.M.; Ungureanu-Iuga, M.; Pogurschi, E.N.; Băbeanu, N.E. Transforming Rhodotorula Sp. Biomass to Active Biologic Compounds for Poultry Nutrition. Agriculture 2023, 13, 1159. [Google Scholar] [CrossRef]
- Ge, Y.; Huang, K.; Xie, W.; Xu, C.; Yao, Q.; Liu, Y. Effects of Rhodotorula mucilaginosa on the Immune Function and Gut Microbiota of Mice. Front. Fungal Biol. 2021, 2, 705696. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Niu, H.; Hua, M.; Su, Y.; Miao, X.; Chi, Y.; Xu, H.; Wang, J.; Sun, M.; et al. Study on the Fermentation Effect of Rhodotorula Glutinis Utilizing Tofu Whey Wastewater and the Influence of Rhodotorula Glutinis on Laying Hens. Front. Nutr. 2023, 10, 1125720. [Google Scholar] [CrossRef]


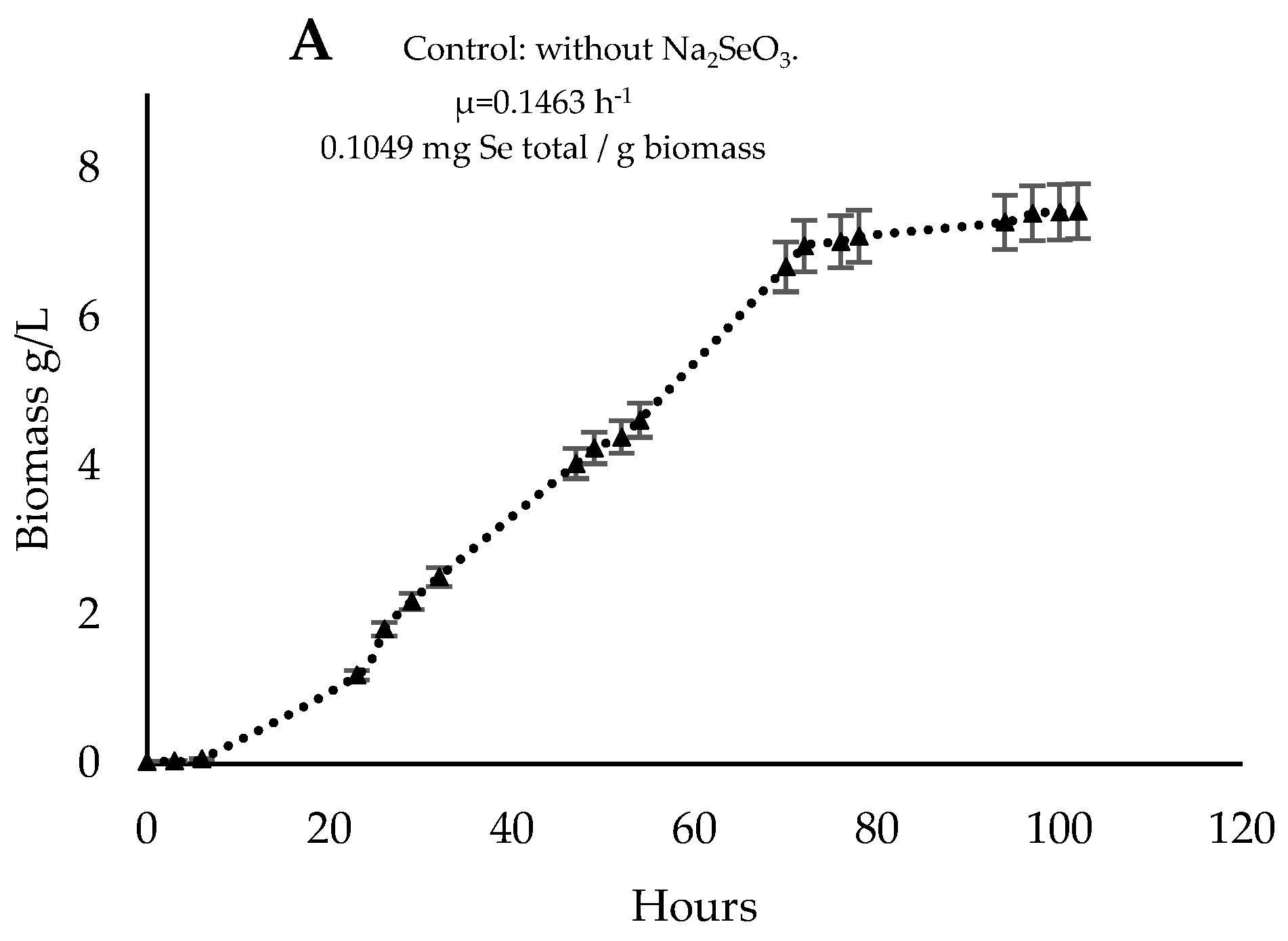
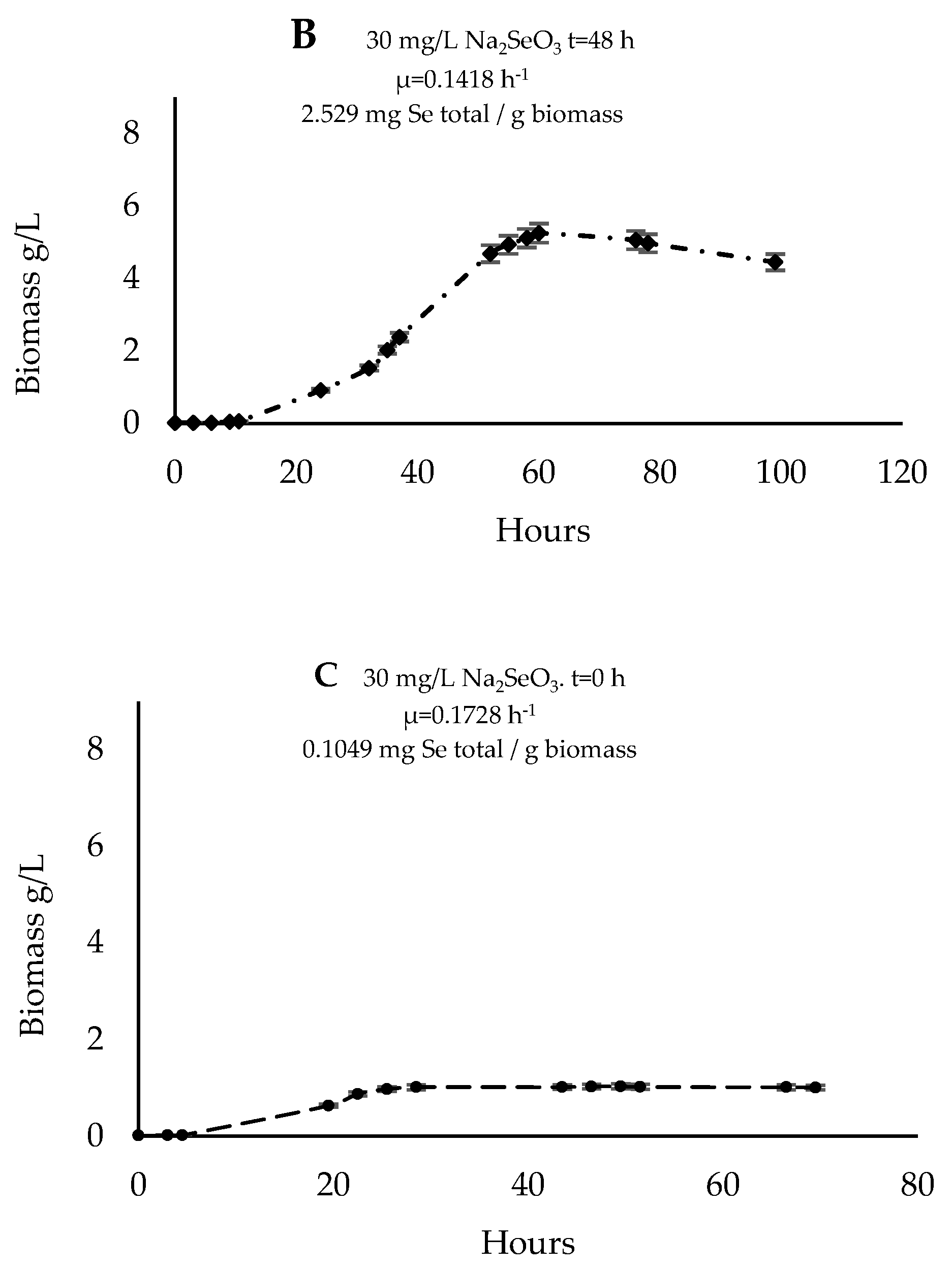
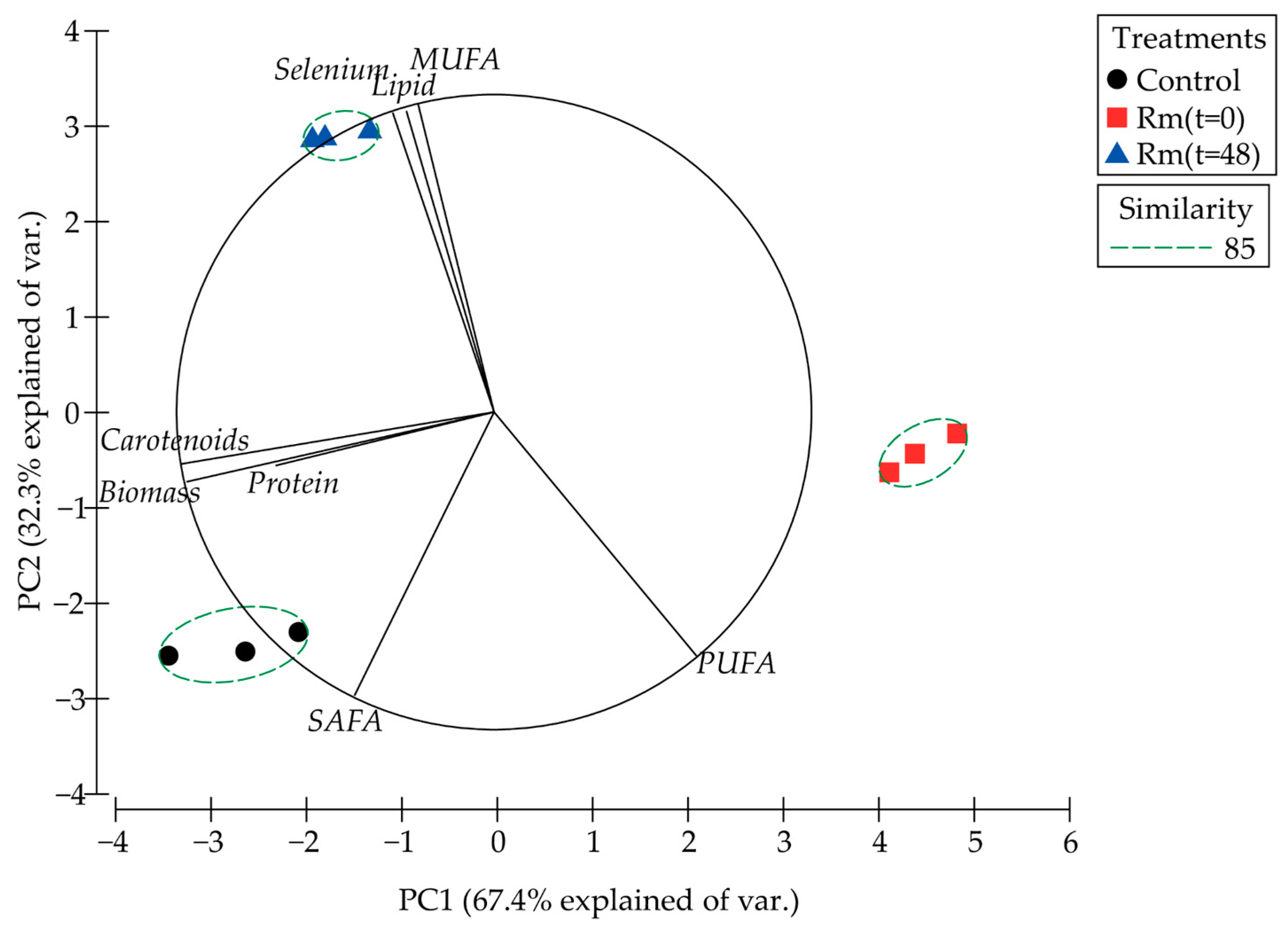
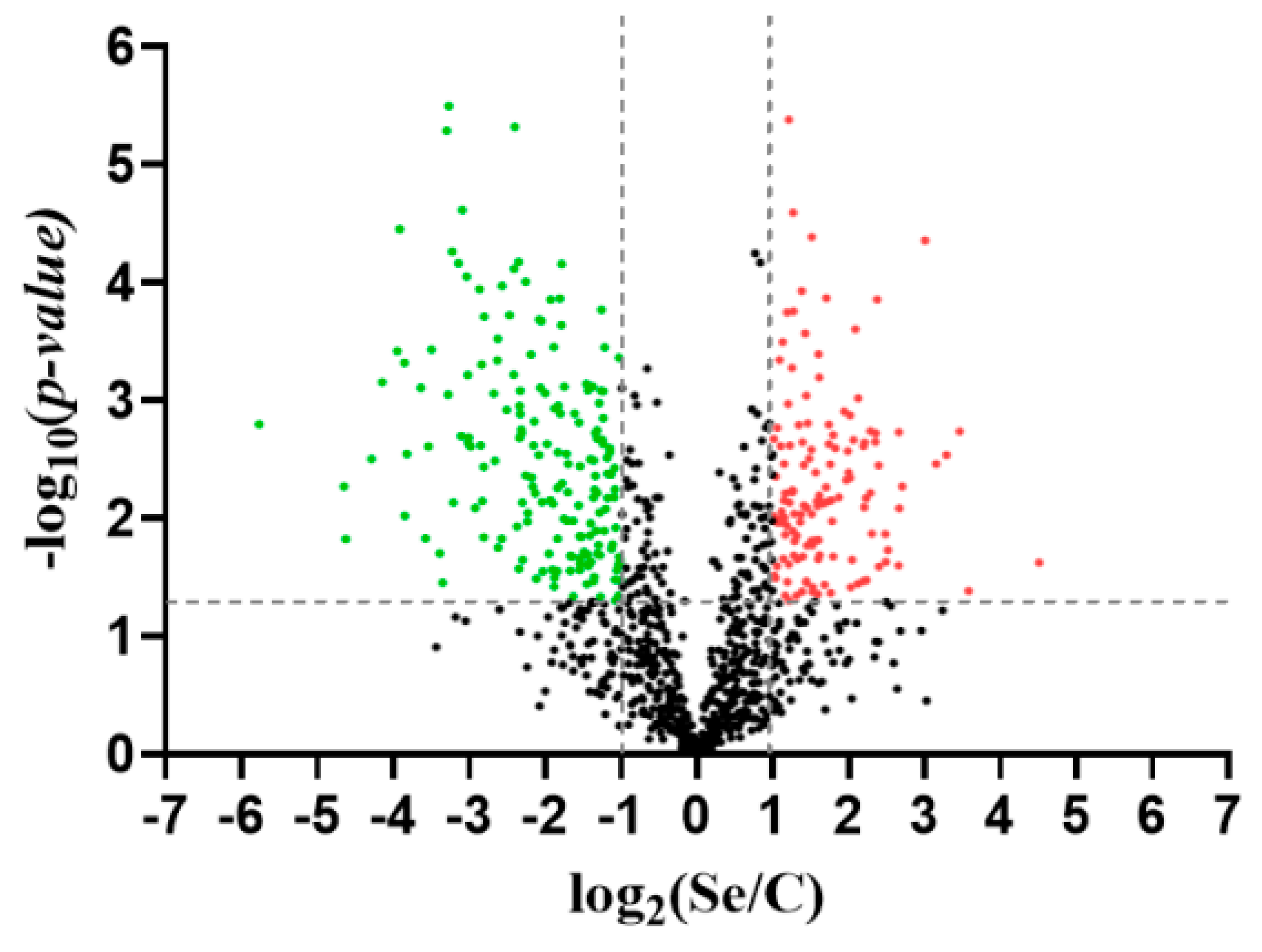
| Parameters | Control | Rm (t = 0) | Rm (t = 48) |
|---|---|---|---|
| Biomass (g/L) | 7.4 ± 0.0 | 1.0 ± 0.1 * | 4.5 ± 0.1 * |
| Total Lipid (%) | 3.2 ± 0.1 | 3.4 ± 0.1 | 4.5 ± 0.0 * |
| SAFA (%) | 47.6 ± 0.4 | 26.9 ± 1.5 * | 22.4 ± 0.5 * |
| MUFA (%) | 24.0 ± 1.1 | 29.6 ± 0.9 * | 69.5 ± 0.6 * |
| PUFA (%) | 29.6 ± 0.5 | 43.6 ± 0.3 * | 8.2 ± 0.2 * |
| Protein (%) | 18.2 ± 0.5 | 16.9 ± 1.2 | 17.7 ± 0.7 |
| Carotenoids (µg/g biomass) | 221.3 ± 22.6 | 63.3 ± 6.0 * | 163.3 ± 8.3 * |
| Selenium (mg/g biomass) | 0.1 ± 0.0 | 0.2 ± 0.0 | 2.5 ± 0.4 * |
| Parameters | Treatments | Selenium |
|---|---|---|
| Biomass (g/L) | −0.462 | 0.022 |
| Total Lipid (%) | 0.902 * | 0.980 * |
| SAFA (%) | −0.935 * | −0.647 |
| MUFA (%) | 0.916 * | 0.983 * |
| PUFA (%) | −0.599 | −0.900 * |
| Protein (%) | −0.227 | 0.108 |
| Carotenoids (µg/g biomass) | −0.357 | 0.131 |
| Selenium (mg/g biomass) | 0.869 * | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Navarrete, P.; Sáez-Arteaga, A.; Marileo, L.; Alors, D.; Correa-Galeote, D.; Dantagnan, P. Enhancing Selenium Accumulation in Rhodotorula mucilaginosa Strain 6S Using a Proteomic Approach for Aquafeed Development. Biomolecules 2024, 14, 629. https://doi.org/10.3390/biom14060629
Díaz-Navarrete P, Sáez-Arteaga A, Marileo L, Alors D, Correa-Galeote D, Dantagnan P. Enhancing Selenium Accumulation in Rhodotorula mucilaginosa Strain 6S Using a Proteomic Approach for Aquafeed Development. Biomolecules. 2024; 14(6):629. https://doi.org/10.3390/biom14060629
Chicago/Turabian StyleDíaz-Navarrete, Paola, Alberto Sáez-Arteaga, Luis Marileo, David Alors, David Correa-Galeote, and Patricio Dantagnan. 2024. "Enhancing Selenium Accumulation in Rhodotorula mucilaginosa Strain 6S Using a Proteomic Approach for Aquafeed Development" Biomolecules 14, no. 6: 629. https://doi.org/10.3390/biom14060629





