Duckweed: Beyond an Efficient Plant Model System
Abstract
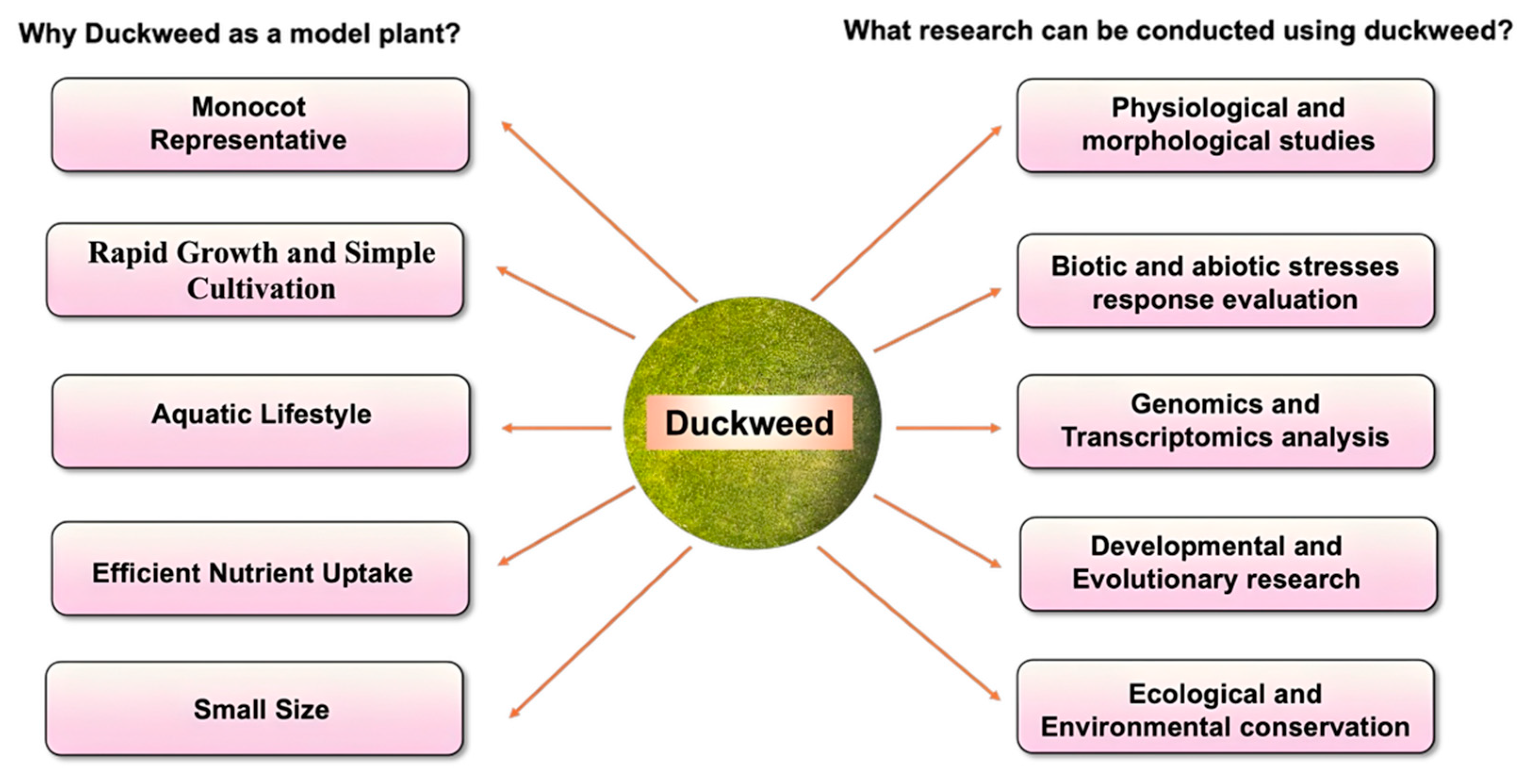
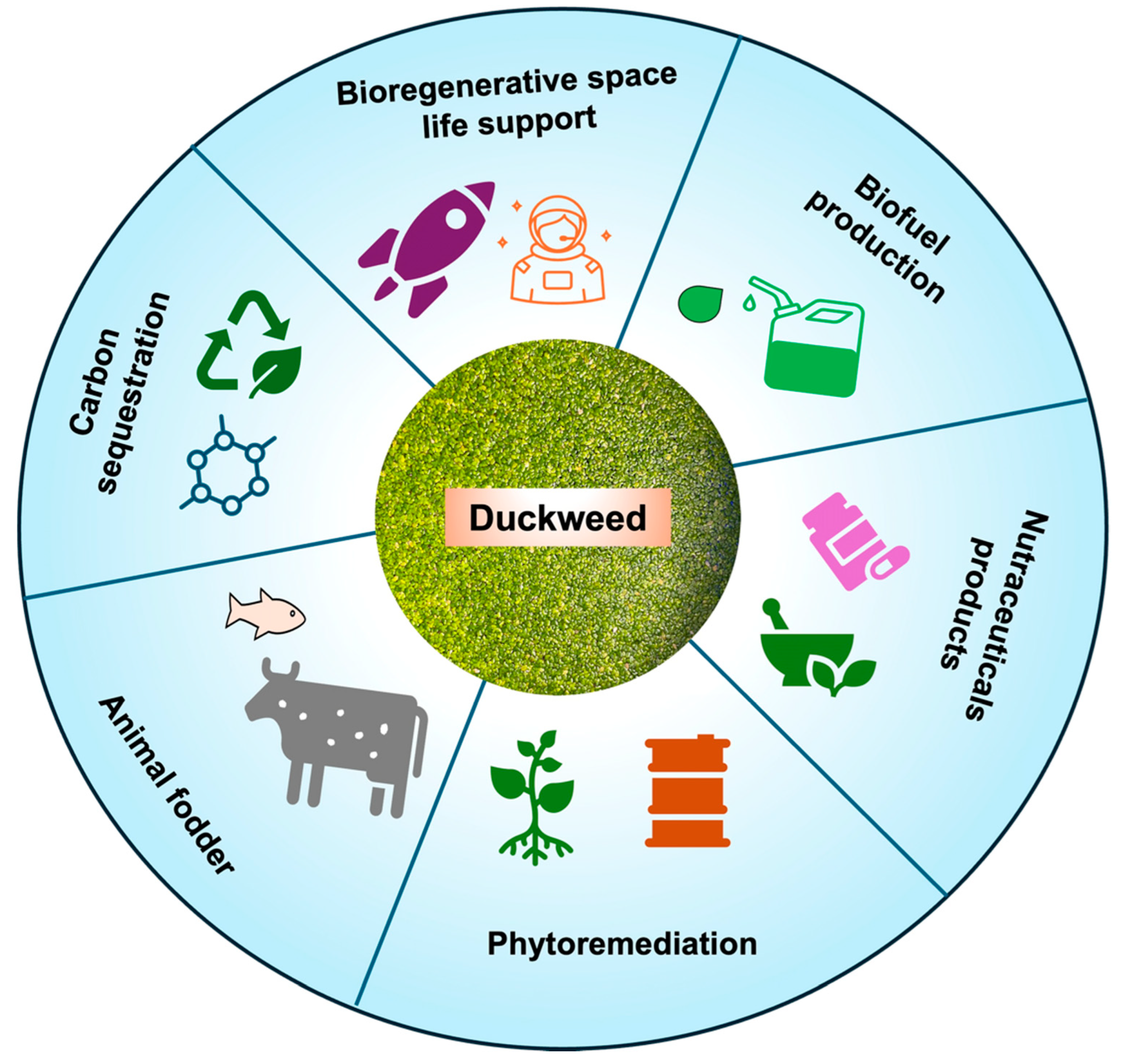
1. Introduction
2. Advantages of Duckweed over Arabidopsis
3. Importance of Duckweed in Physiological Research
3.1. Nutrient Uptake and Stress Tolerance
3.2. Ion Transport in Duckweed
3.3. Signaling Mechanisms in Duckweed
4. Phytoremediation and Wastewater Treatment through Duckweed
5. Genome Complexity of Lemnaceae Species
6. Transgenic Development of Duckweed
7. Development of Novel Methods for Duckweed Germplasm Conservation
8. Discussion and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Landolt, E.; Kandeler, R. The Family of Lemnaceae—A Monographic Study: Phytochemistry, Physiology, Application, and Bibliography; Geobotanical Institute of the ETH, Stiftung Rubel: Zurich, Switzerland, 1986; Volume 1. [Google Scholar]
- Landolt, E.; Kandeler, R. The Family of Lemnaceae—A Monographic Study: Phytochemistry, Physiology, Application, and Bibliography; Geobotanical Institute of the ETH, Stiftung Rubel: Zurich, Switzerland, 1987; Volume 2. [Google Scholar]
- Ziegler, P.; Appenroth, K.J.; Sree, K.S. Survival strategies of duckweeds, the world’s smallest Angiosperms. Plants 2023, 12, 2215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Stepanenko, A.; Kishchenko, O.; Xu, J.; Borisjuk, N. Duckweeds for phytoremediation of polluted water. Plants 2023, 12, 589. [Google Scholar] [CrossRef] [PubMed]
- Baek, G.; Saeed, M.; Choi, H.K. Duckweeds: Their utilization, metabolites and cultivation. Appl. Biol. Chem. 2021, 64, 73. [Google Scholar] [CrossRef] [PubMed]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. Plant Cell 2021, 33, 3207–3234. [Google Scholar] [CrossRef] [PubMed]
- Lasky, J.R.; Des Marais, D.L.; McKay, J.K.; Richards, J.H.; Juenger, T.E.; Keitt, T.H. Characterizing genomic variation of Arabidopsis thaliana: The roles of geography and climate. Mol. Ecol. 2012, 21, 5512–5529. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Y.H.; Su, F.; Chen, L.; Huang, T.; Cai, Y.D. A shortest-path-based method for the analysis and prediction of fruit-related genes in Arabidopsis thaliana. PLoS ONE 2016, 11, e0159519. [Google Scholar] [CrossRef] [PubMed]
- Davila Olivas, N.H.; Frago, E.; Thoen, M.P.; Kloth, K.J.; Becker, F.F.; Van Loon, J.J.; Gort, G.; Keurentjes, J.J.; van Heerwaarden, J.; Dicke, M. Natural variation in life history strategy of Arabidopsis thaliana determines stress responses to drought and insects of different feeding guilds. Mol. Ecol. 2017, 26, 2959–2977. [Google Scholar] [CrossRef] [PubMed]
- Hejný, S.E. Landolt & R. Kandeler The family of Lemnaceae—A monographic study. Folia Geobot. Phytotax 1992, 27, 336. [Google Scholar] [CrossRef]
- Laird, R.A.; Barks, P.M. Skimming the surface: Duckweed as a model system in ecology and evolution. Am. J. Bot. 2018, 105, 1962–1966. [Google Scholar] [CrossRef]
- Iqbal, J.; Javed, A.; Baig, M.A. Growth and nutrient removal efficiency of duckweed (Lemna minor) from synthetic and dumpsite leachate under artificial and natural conditions. PLoS ONE 2019, 14, e0221755. [Google Scholar] [CrossRef]
- Stewart, J.J.; Adams, W.W., III; Escobar, C.M.; López-Pozo, M.; Demmig-Adams, B. Growth and essential carotenoid micronutrients in Lemna gibba as a function of growth light intensity. Front. Plant Sci. 2020, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, P.; Sree, K.S.; Appenroth, K.J. Duckweeds for water remediation and toxicity testing. Toxicol. Environ. Chem. 2016, 98, 1127–1154. [Google Scholar] [CrossRef]
- Ullah, H.; Gul, B.; Khan, H.; Zeb, U. Effect of salt stress on proximate composition of duckweed (Lemna minor L.). Heliyon 2021, 7, e07399. [Google Scholar] [CrossRef]
- Ren, Q.; Xu, Z.; Xue, Y.; Yang, R.; Ma, X.; Sun, J.; Wang, J.; Lin, S.; Wang, W.; Yang, L.; et al. Mechanism of calcium signal response to cadmium stress in duckweed. Plant Signal. Behav. 2022, 17, 2119340. [Google Scholar] [CrossRef] [PubMed]
- Sree, K.S.; Keresztes, Á.; Mueller-Roeber, B.; Brandt, R.; Eberius, M.; Fischer, W.; Appenroth, K.J. Phytotoxicity of cobalt ions on the duckweed Lemna minor–Morphology, ion uptake, and starch accumulation. Chemosphere 2015, 131, 149–156. [Google Scholar] [CrossRef]
- Tian, X.; Fang, Y.; Jin, Y.; Yi, Z.; Li, J.; Du, A.; He, K.; Huang, Y.; Zhao, H. Ammonium detoxification mechanism of ammonium-tolerant duckweed (Landoltia punctata) revealed by carbon and nitrogen metabolism under ammonium stress. Environ. Pollut. 2021, 277, 116834. [Google Scholar] [CrossRef] [PubMed]
- Appenroth, K.J.; Ziegler, P.; Sree, K.S. Accumulation of starch in duckweeds (Lemnaceae), potential energy plants. Physiol. Mol. Biol. Plants 2021, 27, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Wang, X.; Fang, Y.; Zhang, Y.; Huang, M.; Zhao, H. The influence of different plant hormones on biomass and starch accumulation of duckweed: A renewable feedstock for bioethanol production. Renew. Energy 2019, 138, 659–665. [Google Scholar] [CrossRef]
- Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin-induced growth in the duckweeds Lemna gibba and Spirodela polyrhiza. Plant Growth Regul. 2018, 86, 477–486. [Google Scholar] [CrossRef]
- Yang, L.; Sun, J.; Yan, C.; Wu, J.; Wang, Y.; Ren, Q.; Wang, S.; Ma, X.; Zhao, L.; Sun, J. Regeneration of duckweed (Lemna turonifera) involves genetic molecular regulation and cyclohexane release. PLoS ONE 2022, 17, e0254265. [Google Scholar] [CrossRef]
- Huang, M.; Fang, Y.; Liu, Y.; Jin, Y.; Sun, J.; Tao, X.; Ma, X.; He, K.; Zhao, H. Using proteomic analysis to investigate uniconazole-induced phytohormone variation and starch accumulation in duckweed (Landoltia punctata). BMC Biotechnol. 2015, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Fang, Y.; Xiao, Y.; Jin, Y.L.; Ma, X.R.; Zhao, Y.; He, K.Z.; Zhao, H.; Wang, H.Y. Comparative transcriptome analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Biotechnol. Biofuels 2013, 6, 72. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Ding, Z.; Sun, X.; Zhang, J. Physiological and transcriptomic analysis reveals distorted ion homeostasis and responses in the freshwater plant Spirodela polyrhiza L. under salt stress. Genes 2019, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yao, J.; Sun, J.; Shi, L.; Chen, Y.; Sun, J. The Ca2+ signaling, Glu, and GABA responds to Cd stress in duckweed. Aquat. Toxicol. 2020, 218, 105352. [Google Scholar] [CrossRef] [PubMed]
- Razinger, J.; Dermastia, M.; Koce, J.D.; Zrimec, A. Oxidative stress in duckweed (Lemna minor L.) caused by short-term cadmium exposure. Environ. Pollut. 2008, 153, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Baggs, E.L.; Tiersma, M.B.; Abramson, B.W.; Michael, T.P.; Krasileva, K.V. Characterization of defense responses against bacterial pathogens in duckweeds lacking EDS1. New Phytol. 2022, 236, 1838–1855. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Taoka, K.I.; Hosaka, A.; Tanaka, K.; Kobayashi, H.; Muranaka, T.; Toyooka, K.; Oyama, T.; Tsuji, H. Characterization of frond and flower development and identification of FT and FD genes from duckweed Lemna aequinoctialis Nd. Front. Plant Sci. 2021, 12, 697206. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yu, C.; Xia, X.; Li, M.; Li, H.; Wang, Y.; Wang, S.; Wang, C.; Ma, Y.; Zhou, G. Comparative transcriptome analysis of duckweed (Landoltia punctata) in response to cadmium provides insights into molecular mechanisms underlying hyperaccumulation. Chemosphere 2018, 190, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Fang, Y.; Huang, J.; Zhao, Y.; Li, Q.; Lai, F.; Xu, Y.; Tian, X.; He, K.; Jin, Y.; et al. Duckweed systems for eutrophic water purification through converting wastewater nutrients to high-starch biomass: Comparative evaluation of three different genera (Spirodela polyrhiza, Lemna minor and Landoltia punctata) in monoculture or polyculture. RSC Adv. 2023, 8, 17927–17937. [Google Scholar] [CrossRef]
- Körner, S.; Vermaat, J.E.; Veenstra, S. The capacity of duckweed to treat wastewater: Ecological considerations for a sound design. J. Environ. Qual. 2003, 32, 1583–1590. [Google Scholar] [CrossRef]
- Ubuza, L.J.A.; Padero, P.C.S.; Nacalaban, C.M.N.; Tolentino, J.T.; Alcoran, D.C.; Tolentino, J.C.; Ido, A.L.; Mabayo, V.I.F.; Arazo, R.O. Assessment of the potential of duckweed (Lemna minor L.) in treating lead-contaminated water through phytoremediation in stationary and recirculated set-ups. Environ. Eng. Res. 2020, 25, 977–982. [Google Scholar] [CrossRef]
- Ahmadi, A.W.; Dursun, S. Assessing the Efficiency and Role of Duckweed (Lemna Minor) in the Removal of Pollutants from Wastewater Treatment Plant Secondary Clarifier Tanks: A Comprehensive Review. Cent. Asian J. Water Res. 2024, 10, 115–125. [Google Scholar] [CrossRef]
- Liu, Z.; Tran, K.Q. A review on disposal and utilization of phytoremediation plants containing heavy metals. Ecotoxicol. Environ. Saf. 2021, 226, 112821. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Maiti, S.K. Biochar assisted phytoremediation and biomass disposal in heavy metal contaminated mine soils: A review. Int. J. Phytoremediation 2021, 23, 559–576. [Google Scholar] [CrossRef] [PubMed]
- Hazotte, C.; Laubie, B.; Rees, F.; Morel, J.L.; Simonnot, M.O. A novel process to recover cadmium and zinc from the hyperaccumulator plant Noccaea caerulescens. Hydrometallurgy 2017, 174, 56–65. [Google Scholar] [CrossRef]
- Barbaroux, R.; Mercier, G.; Blais, J.F.; Morel, J.L.; Simonnot, M.O. A new method for obtaining nickel metal from the hyperaccumulator plant Alyssum murale. Sep. Purif. Technol. 2011, 83, 57–65. [Google Scholar] [CrossRef]
- Tang, M.D.; Wu, L.H.; Li, N.; Luo, Y.M.; Hu, F.; Zhang, L.X. Preliminary study on effect of Elsholtzia splendens compost on plant growth and Cu uptake by winter wheat in a Cu-deficient upland soil. Soils 2006, 38, 614–618. [Google Scholar]
- Cao, Z.; Wang, S.; Wang, T.; Chang, Z.; Shen, Z.; Chen, Y. Using contaminated plants involved in phytoremediation for anaerobic digestion. Int. J. Phytoremediation 2015, 17, 201–207. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Li, C.; Zhou, Y.; Wu, Y.; Wang, W. Genomes and transcriptomes of duckweeds. Front. Chem. 2018, 6, 230. [Google Scholar] [CrossRef]
- Wang, W.; Li, R.; Zhu, Q.; Tang, X.; Zhao, Q. Transcriptomic and physiological analysis of common duckweed Lemna minor responses to NH4+ toxicity. BMC Plant Biol. 2016, 16, 92. [Google Scholar] [CrossRef]
- Harkess, A.; Bewick, A.J.; Lu, Z.; Fourounjian, P.; Michael, T.P.; Schmitz, R.J.; Meyers, B.C. The unusual predominance of maintenance DNA methylation in Spirodela polyrhiza. G3 Genes Genomes Genet. 2024, 14, jkae004. [Google Scholar] [CrossRef] [PubMed]
- Van Hoeck, A.; Horemans, N.; Monsieurs, P.; Cao, H.X.; Vandenhove, H.; Blust, R. The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol. Biofuels 2015, 8, 188. [Google Scholar] [CrossRef] [PubMed]
- Hoang, P.T.; Fiebig, A.; Novák, P.; Macas, J.; Cao, H.X.; Stepanenko, A.; Chen, G.; Borisjuk, N.; Scholz, U.; Schubert, I. Chromosome-scale genome assembly for the duckweed Spirodela intermedia, integrating cytogenetic maps, PacBio and Oxford Nanopore libraries. Sci. Rep. 2020, 10, 19230. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Haberer, G.; Gundlach, H.; Gläßer, C.; Nussbaumer, T.C.L.M.; Luo, M.C.; Lomsadze, A.; Borodovsky, M.; Kerstetter, R.A.; Shanklin, J.; et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014, 5, 3311. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Park, J.H.; Lee, Y.; Woo, D.U.; Jeon, H.H.; Sung, Y.W.; Shim, S.; Kim, S.H.; Lee, K.O.; Kim, J.Y.; et al. Genome of the world’s smallest flowering plant, Wolffia australiana, helps explain its specialized physiology and unique morphology. Commun. Biol. 2021, 4, 900. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P.; VanBuren, R. Progress, challenges and the future of crop genomes. Curr. Opin. Plant Biol. 2015, 24, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Schatz, M.C.; Delcher, A.L.; Salzberg, S.L. Assembly of large genomes using second-generation sequencing. Genome Res. 2010, 20, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Michael, T.P.; Bryant, D.; Gutierrez, R.; Borisjuk, N.; Chu, P.; Zhang, H.; Xia, J.; Zhou, J.; Peng, H.; El Baidouri, M.; et al. Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 2017, 89, 617–635. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Gnerre, S.; MacCallum, I.; Przybylski, D.; Ribeiro, F.J.; Burton, J.N.; Walker, B.J.; Sharpe, T.; Hall, G.; Shea, T.P.; Sykes, S.; et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 2011, 108, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The third revolution in sequencing technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, D.; Mathur, S.; Singh, A.; Ranjan, R. Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 2022, 13, 1030890. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinforma 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Schöpflin, R.; Melo, U.S.; Moeinzadeh, H.; Heller, D.; Laupert, V.; Hertzberg, J.; Holtgrewe, M.; Alavi, N.; Klever, M.K.; Jungnitsch, J.; et al. Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes. Nat. Commun. 2022, 13, 6470. [Google Scholar] [CrossRef]
- Belton, J.M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef]
- Yamamoto, Y.T.; Rajbhandari, N.; Lin, X.; Bergmann, B.A.; Nishimura, Y.; Stomp, A.M. Genetic transformation of duckweed Lemna gibba and Lemna minor. Vitr. Cell. Dev. Biol.-Plant 2001, 37, 349–353. [Google Scholar] [CrossRef]
- Ko, S.M.; Sun, H.J.; Oh, M.J.; Song, I.J.; Kim, M.J.; Sin, H.S.; Goh, C.H.; Kim, Y.W.; Lim, P.O.; Lee, H.Y.; et al. Expression of the protective antigen for PEDV in transgenic duckweed, Lemna minor. Hortic. Environ. Biotechnol. 2011, 52, 511–515. [Google Scholar] [CrossRef]
- Firsov, A.; Tarasenko, I.; Mitiouchkina, T.; Shaloiko, L.; Kozlov, O.; Vinokurov, L.; Rasskazova, E.; Murashev, A.; Vainstein, A.; Dolgov, S. Expression and immunogenicity of M2e peptide of avian influenza virus H5N1 fused to ricin toxin b chain produced in duckweed plants. Front. Chem. 2018, 6, 22. [Google Scholar] [CrossRef]
- Tan, X.; Chen, S.; Fang, Y.; Liu, P.; Hu, Z.; Jin, Y.; Yi, Z.; He, K.; Li, X.; Zhao, L.; et al. Rapid and highly efficient genetic transformation and application of interleukin-17B expressed in duckweed as mucosal vaccine adjuvant. Biomolecules 2022, 12, 1881. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, X.H.; Anaokar, S.; Shi, H.; Dahl, W.B.; Cai, Y.; Luo, G.; Chai, J.; Cai, Y.; Mollá-Morales, A.; et al. Engineering triacylglycerol accumulation in duckweed (Lemna japonica). Plant Biotechnol. J. 2023, 21, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cheng, J.J.; Himmel, M.E.; Skory, C.D.; Adney, W.S.; Thomas, S.R.; Tisserat, B.; Nishimura, Y.; Yamamoto, Y.T. Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresour. Technol. 2007, 98, 2866–2872. [Google Scholar] [CrossRef]
- Yang, L.; Han, H.; Liu, M.; Zuo, Z.; Zhou, K.; Lü, J.; Zhu, Y.; Bai, Y.; Wang, Y. Overexpression of the Arabidopsis photorespiratory pathway gene, serine: Glyoxylate aminotransferase (AtAGT1), leads to salt stress tolerance in transgenic duckweed (Lemna minor). Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 407–416. [Google Scholar] [CrossRef]
- Yang, L.; Wei, Y.; Li, N.; Zeng, J.; Han, Y.; Zuo, Z.; Wang, S.; Zhu, Y.; Zhang, Y.; Sun, J.; et al. Declined cadmium accumulation in Na+/H+ antiporter (NHX1) transgenic duckweed under cadmium stress. Ecotoxicol. Environ. Saf. 2019, 182, 109397. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Xu, S.; Tang, X.; Zhao, J.; Yu, C.; He, G.; Xu, H.; Wang, S.; Tang, Y.; et al. Efficient genetic transformation and CRISPR/Cas9-mediated genome editing in Lemna aequinoctialis. Plant Biotechnol. J. 2019, 17, 2143–2152. [Google Scholar] [CrossRef]
- Peterson, A.; Kishchenko, O.; Kuhlmann, M.; Tschiersch, H.; Fuchs, J.; Tikhenko, N.; Schubert, I.; Nagel, M. Cryopreservation of Duckweed Genetic Diversity as Model for Long-Term Preservation of Aquatic Flowering Plants. Plants 2023, 12, 3302. [Google Scholar] [CrossRef] [PubMed]
- N’Nan, O.; Hocher, V.; Verdeil, J.L.; Konan, J.L.; Ballo, K.; Mondeil, F.; Malaurie, B. Cryopreservation by encapsulation–dehydration of plumules of coconut (Cocos nucifera L.). CryoLetters 2008, 29, 339–350. [Google Scholar] [PubMed]
- Palanyandy, S.R.; Gantait, S.; Subramaniam, S.; Sinniah, U.R. Cryopreservation of oil palm (Elaeis guineensis Jacq.) polyembryoids via encapsulation–desiccation. 3 Biotech 2020, 10, 9. [Google Scholar] [CrossRef]
- Parsons, J.L.; Wingate, V. Biolex Therapeutics Inc. Methods and Compositions for the Cryopreservation of Duckweed. U.S. Patent 13/379,959, 28 February 2014. [Google Scholar]
- Appenroth, K.J.; Sree, K.S.; Böhm, V.; Hammann, S.; Vetter, W.; Leiterer, M.; Jahreis, G. Nutritional value of duckweeds (Lemnaceae) as human food. Food Chem. 2017, 217, 266–273. [Google Scholar] [CrossRef]
- Edelman, M.; Colt, M. Nutrient value of leaf vs. seed. Front. Chem. 2016, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, H.; Stomp, A.M.; Cheng, J.J. The production of duckweed as a source of biofuels. Biofuels 2012, 3, 589–601. [Google Scholar] [CrossRef]
- Rival, S.; Wisniewski, J.P.; Langlais, A.; Kaplan, H.; Freyssinet, G.; Vancanneyt, G.; Vunsh, R.; Perl, A.; Edelman, M. Spirodela (duckweed) as an alternative production system for pharmaceuticals: A case study, aprotinin. Transgenic Res. 2008, 17, 503–513. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thingujam, D.; Pajerowska-Mukhtar, K.M.; Mukhtar, M.S. Duckweed: Beyond an Efficient Plant Model System. Biomolecules 2024, 14, 628. https://doi.org/10.3390/biom14060628
Thingujam D, Pajerowska-Mukhtar KM, Mukhtar MS. Duckweed: Beyond an Efficient Plant Model System. Biomolecules. 2024; 14(6):628. https://doi.org/10.3390/biom14060628
Chicago/Turabian StyleThingujam, Doni, Karolina M. Pajerowska-Mukhtar, and M. Shahid Mukhtar. 2024. "Duckweed: Beyond an Efficient Plant Model System" Biomolecules 14, no. 6: 628. https://doi.org/10.3390/biom14060628
APA StyleThingujam, D., Pajerowska-Mukhtar, K. M., & Mukhtar, M. S. (2024). Duckweed: Beyond an Efficient Plant Model System. Biomolecules, 14(6), 628. https://doi.org/10.3390/biom14060628




