Enhancing Selective Antimicrobial and Antibiofilm Activities of Melittin through 6-Aminohexanoic Acid Substitution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Peptide Synthesis
2.3. Circular Dichroism (CD) Spectroscopy
2.4. Antimicrobial Activity
2.5. Hemolytic Activity
2.6. Cytotoxicity against RAW 264.7 Cells
2.7. Membrane Depolarization
2.8. SYTOX Green Uptake Assay
2.9. Membrane Permeability Assay
2.10. Antibiofilm Activity
2.11. Confocal Laser Scanning Microscopy
2.12. FACScan Analysis
3. Results and Discussion
3.1. Peptide Design and Characterization
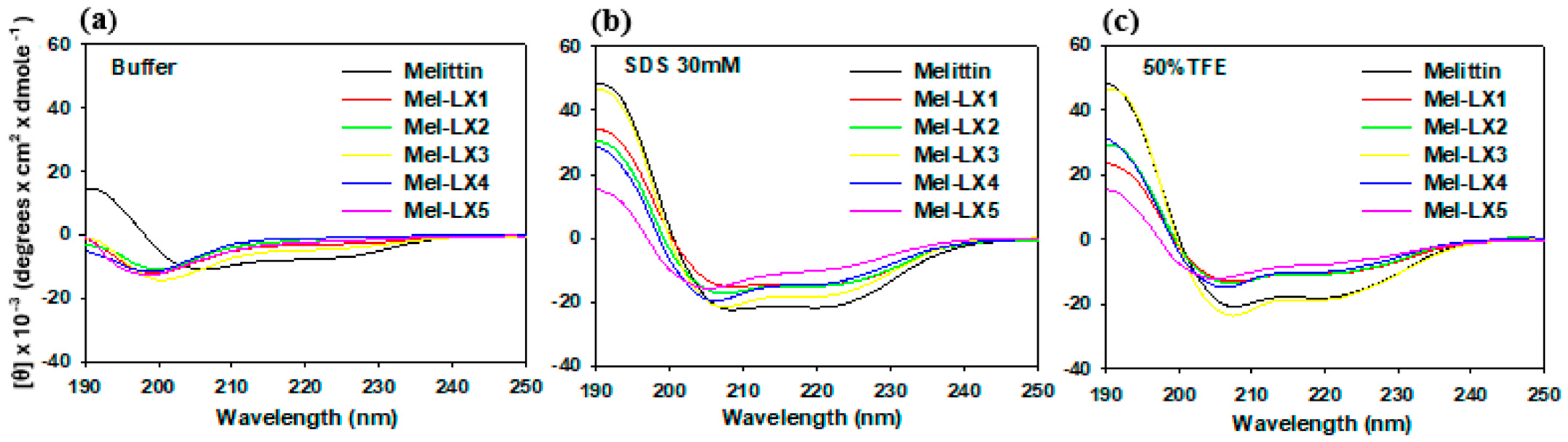
3.2. CD Spectroscopy
3.3. Antibacterial Activity of Melittin and Its Analogs
3.4. Hemolytic and Cytotoxic Activities of Melittin and Its Analogs
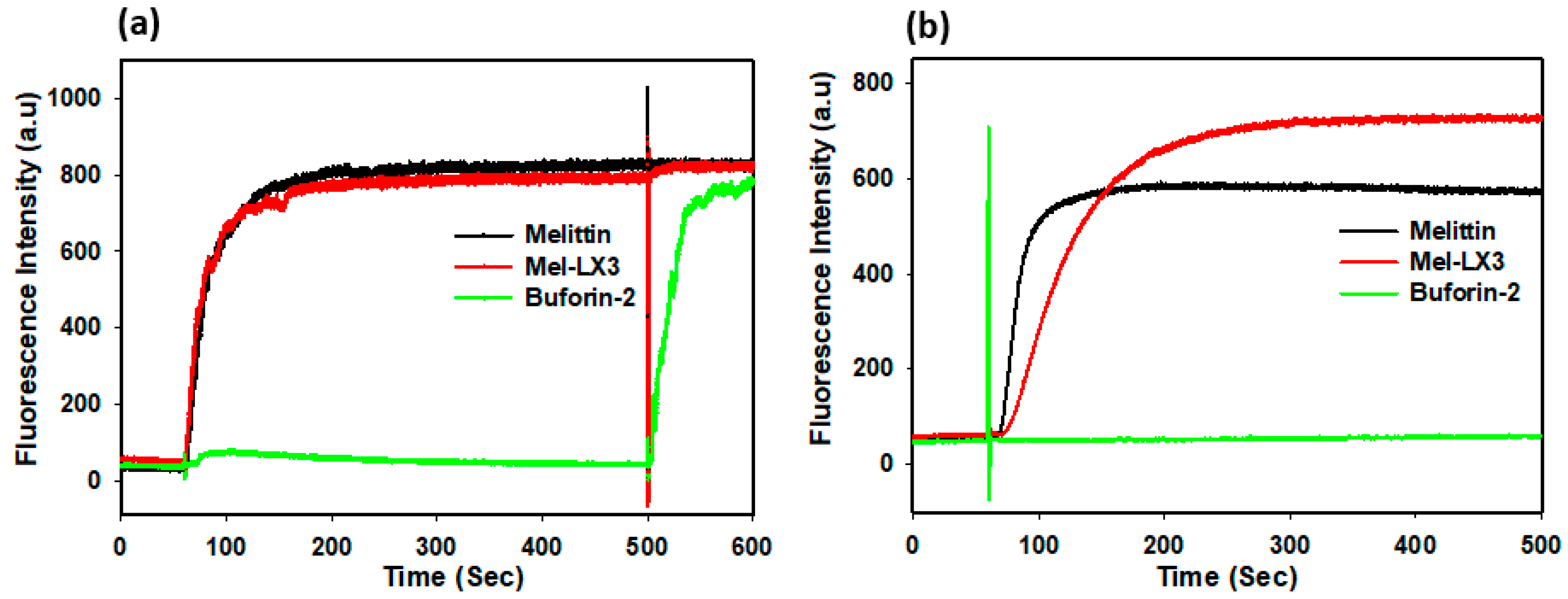
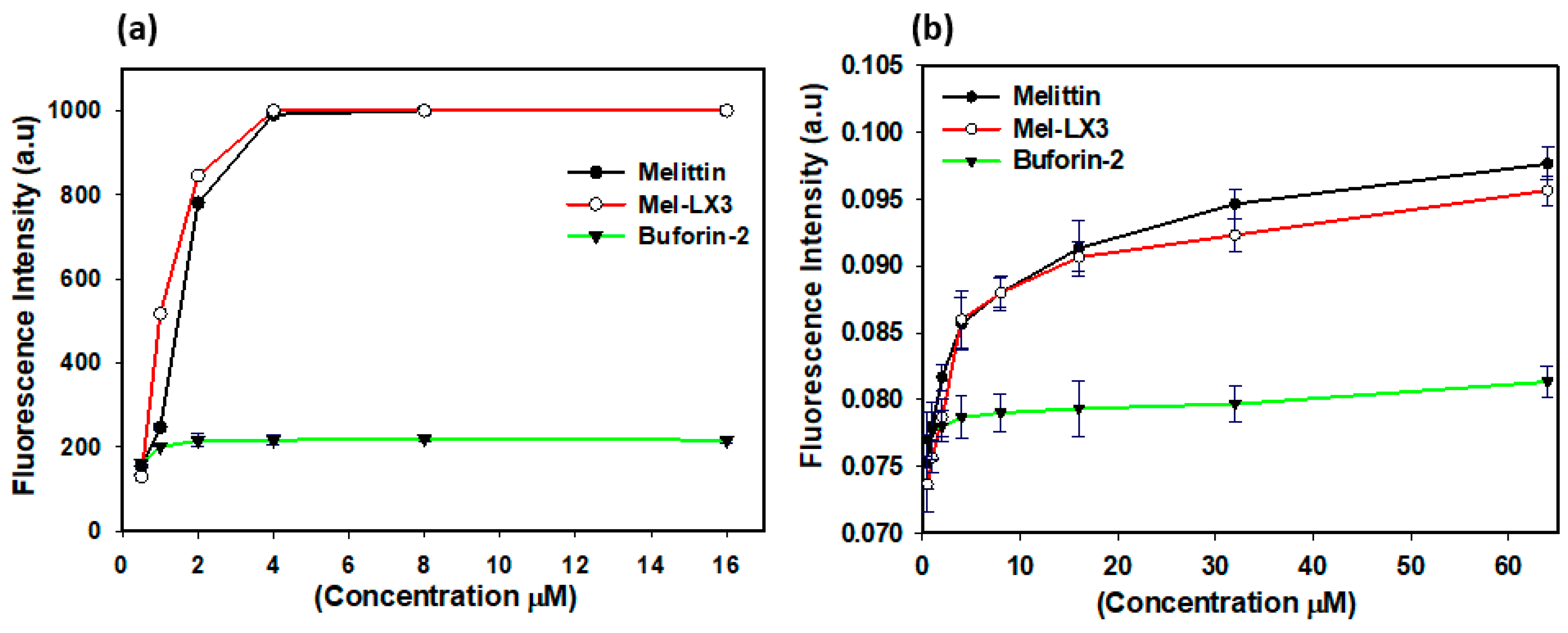
3.5. Membrane Permeabilization of Gram-Positive Bacteria
3.6. Membrane Permeabilization of Gram-Negative Bacteria
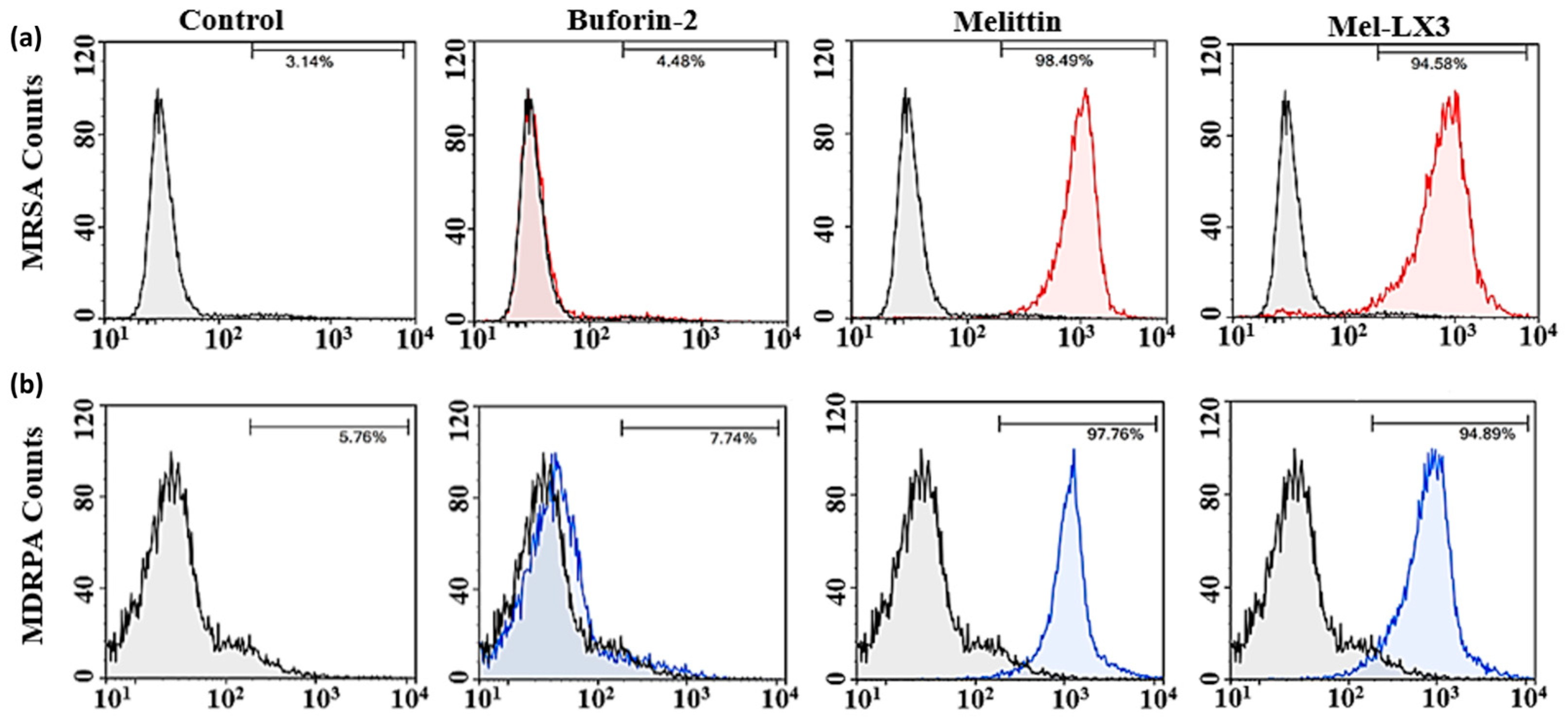
3.7. Flow Cytometric Analysis
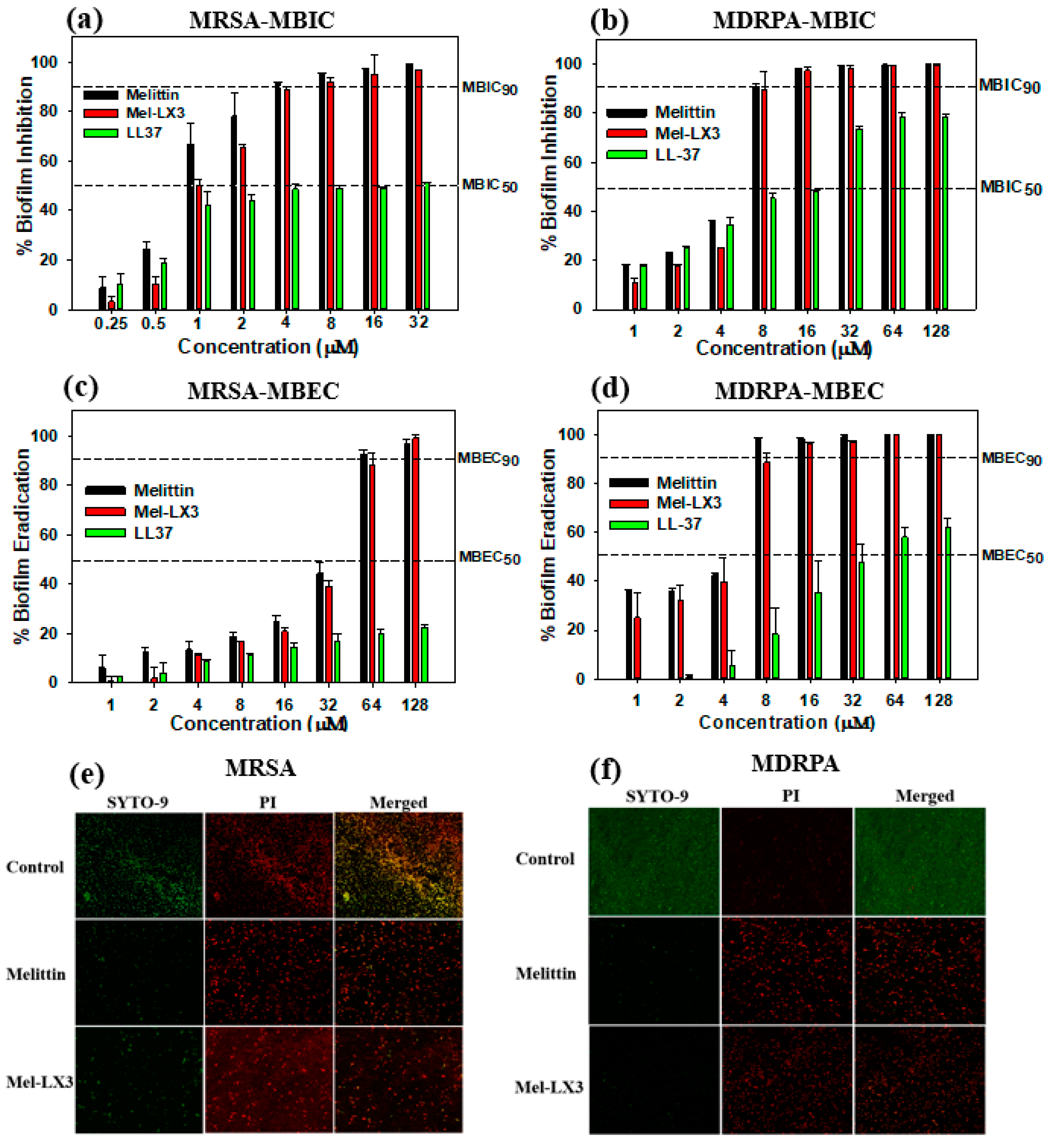
3.8. Antibiofilm Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Yedery, R.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef] [PubMed]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial peptides: Features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Mba, I.E.; Nweze, E.I. Focus: Antimicrobial resistance: Antimicrobial peptides therapy: An emerging alternative for treating drug-resistant bacteria. Yale J. Biol. Med. 2022, 95, 445. [Google Scholar]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial peptides: A new hope in biomedical and pharmaceutical fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Li, W.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. Chemically modified and conjugated antimicrobial peptides against superbugs. Chem. Soc. Rev. 2021, 50, 4932–4973. [Google Scholar] [CrossRef]
- Silva, O.N.; Mulder, K.C.; Barbosa, A.E.; Otero-Gonzalez, A.J.; Lopez-Abarrategui, C.; Rezende, T.M.; Dias, S.C. Exploring the pharmacological potential of promiscuous host-defense peptides: From natural screenings to biotechnological applications. Front. Microbiol. 2011, 2, 232. [Google Scholar]
- Nayab, S.; Aslam, M.A.; Rahman, S.U.; Sindhu, Z.U.D.; Sajid, S.; Zafar, N.; Razaq, M.; Kanwar, R.; Amanullah. A review of antimicrobial peptides: Its function, mode of action and therapeutic potential. Int. J. Pept. Res. Ther. 2022, 28, 46. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef] [PubMed]
- Guha, S.; Ferrie, R.P.; Ghimire, J.; Ventura, C.R.; Wu, E.; Sun, L.; Kim, S.Y.; Wiedman, G.R.; Hristova, K.; Wimley, W.C. Applications and evolution of melittin, the quintessential membrane active peptide. Biochem. Pharmacol. 2021, 193, 114769. [Google Scholar] [CrossRef]
- Lee, M.-T.; Sun, T.-L.; Hung, W.-C.; Huang, H.W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, C.; Cheng, J.; Huang, H.; Lovell, J.F.; Jin, H. Delivery strategies for melittin-based cancer therapy. ACS Appl. Mater. Interfaces 2021, 13, 17158–17173. [Google Scholar] [CrossRef]
- Lee, J.A.; Son, M.J.; Choi, J.; Jun, J.H.; Kim, J.-I.; Lee, M.S. Bee venom acupuncture for rheumatoid arthritis: A systematic review of randomised clinical trials. BMJ Open 2014, 4, e006140. [Google Scholar] [CrossRef]
- Jeong, Y.-J.; Shin, J.-M.; Bae, Y.-S.; Cho, H.-J.; Park, K.-K.; Choe, J.-Y.; Han, S.M.; Moon, S.K.; Kim, W.J.; Choi, Y.H.; et al. Melittin has a chondroprotective effect by inhibiting MMP-1 and MMP-8 expressions via blocking NF-κB and AP-1 signaling pathway in chondrocytes. Int. Immunopharmacol. 2015, 25, 400–405. [Google Scholar] [CrossRef]
- Son, D.J.; Lee, J.W.; Lee, Y.H.; Song, H.S.; Lee, C.K.; Hong, J.T. Therapeutic application of anti-arthritis, pain-releasing, and anti-cancer effects of bee venom and its constituent compounds. Pharmacol. Ther. 2007, 115, 246–270. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Kim, M.J.; Yoon, I.; Li, D.X.; Bae, H.; Kim, S.K. Nicotinic acetylcholine receptors mediate the suppressive effect of an injection of diluted bee venom into the GV3 acupoint on oxaliplatin-induced neuropathic cold allodynia in rats. Biol. Pharm. Bull. 2015, 38, 710–714. [Google Scholar] [CrossRef]
- Lim, B.-S.; Moon, H.J.; Li, D.X.; Gil, M.; Min, J.K.; Lee, G.; Bae, H.; Kim, S.K.; Min, B.-I. Effect of bee venom acupuncture on oxaliplatin-induced cold allodynia in rats. Evid.-Based Complement. Altern. Med. 2013, 2013, 369324. [Google Scholar] [CrossRef]
- Varanda, E.A.; Monti, R.; Tavares, D.C. Inhibitory effect of propolis and bee venom on the mutagenicity of some direct-and indirect-acting mutagens. Teratog. Carcinog. Mutagen. 1999, 19, 403–413. [Google Scholar] [CrossRef]
- Hu, H.; Chen, D.; Li, Y.; Zhang, X. Effect of polypeptides in bee venom on growth inhibition and apoptosis induction of the human hepatoma cell line SMMC-7721 in-vitro and Balb/c nude mice in-vivo. J. Pharm. Pharmacol. 2006, 58, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.-E.; Baek, Y.-H.; Lee, M.-H.; Choi, D.-Y.; Park, D.-S.; Lee, J.-D. Bee venom inhibits tumor angiogenesis and metastasis by inhibiting tyrosine phosphorylation of VEGFR-2 in LLC-tumor-bearing mice. Cancer Lett. 2010, 292, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Gajski, G.; Garaj-Vrhovac, V. Radioprotective effects of honeybee venom (Apis mellifera) against 915-mhz microwave radiation–induced DNA damage in wistar rat lymphocytes: In vitro study. Int. J. Toxicol. 2009, 28, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Benz, R.; Hancock, R.E. Influence of proline residues on the antibacterial and synergistic activities of α-helical peptides. Biochemistry 1999, 38, 8102–8111. [Google Scholar] [CrossRef]
- Lam, Y.; Wassall, S.; Morton, C.; Smith, R.; Separovic, F. Solid-state NMR structure determination of melittin in a lipid environment. Biophys. J. 2001, 81, 2752–2761. [Google Scholar] [CrossRef]
- Lam, Y.-H.; Morton, C.; Separovic, F. Solid-state NMR conformational studies of a melittin-inhibitor complex. Eur. Biophys. J. 2002, 31, 383–388. [Google Scholar]
- Ladokhin, A.S.; Selsted, M.E.; White, S.H. Sizing membrane pores in lipid vesicles by leakage of co-encapsulated markers: Pore formation by melittin. Biophys. J. 1997, 72, 1762–1766. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, R.; Bhunia, A.; Bhattacharjya, S. Micelle-bound structures and dynamics of the hinge deleted analog of melittin and its diastereomer: Implications in cell selective lysis by d-amino acid containing antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 128–139. [Google Scholar] [CrossRef]
- Asthana, N.; Yadav, S.P.; Ghosh, J.K. Dissection of antibacterial and toxic activity of melittin: A leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J. Biol. Chem. 2004, 279, 55042–55050. [Google Scholar] [CrossRef]
- Markowska, A.; Markowski, A.R.; Jarocka-Karpowicz, I. The importance of 6-aminohexanoic acid as a hydrophobic, flexible structural element. Int. J. Mol. Sci. 2021, 22, 12122. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Chen, Y.-H.; Wang, P.; Zhang, J.; Gurewich, V.; Zhang, P.; Liu, J.-N. The blockage of the high-affinity lysine binding sites of plasminogen by EACA significantly inhibits prourokinase-induced plasminogen activation. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2002, 1596, 182–192. [Google Scholar] [CrossRef]
- Story, S.C.; Aldrich, J.V. Preparation of protected peptide amides using the Fmoc chemical protocol: Comparison of resins for solid phase synthesis 1. Int. J. Pept. Protein Res. 1992, 39, 87–92. [Google Scholar] [CrossRef]
- Yang, S.-T.; Shin, S.-Y.; Shin, S.-H. The central PXXP motif is crucial for PMAP-23 translocation across the lipid bilayer. Int. J. Mol. Sci. 2021, 22, 9752. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Shin, S.-H.; Yang, S. Rationally designed PMAP-23 derivatives with enhanced bactericidal and anticancer activity based on the molecular mechanism of peptide–membrane interactions. Amino Acids 2023, 55, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, 10–128. [Google Scholar] [CrossRef] [PubMed]
- Jahan, I.; Kumar, S.D.; Shin, S.Y.; Lee, C.W.; Shin, S.-H.; Yang, S. Multifunctional properties of BMAP-18 and its aliphatic analog against drug-resistant bacteria. Pharmaceuticals 2023, 16, 1356. [Google Scholar] [CrossRef]
- Lee, H.; Lim, S.I.; Shin, S.-H.; Lim, Y.; Koh, J.W.; Yang, S. Conjugation of cell-penetrating peptides to antimicrobial peptides enhances antibacterial activity. ACS Omega 2019, 4, 15694–15701. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lee, C.W.; Kim, H.J.; Jung, H.-H.; Kim, J.I.; Shin, S.Y.; Shin, S.-H. Structural analysis and mode of action of BMAP-27, a cathelicidin-derived antimicrobial peptide. Peptides 2019, 118, 170106. [Google Scholar] [CrossRef]
- Lee, H.; Yang, S.; Shin, S.-H. Effect of central PxxP motif in amphipathic alpha-helical peptides on antimicrobial activity and mode of action. J. Anal. Sci. Technol. 2023, 14, 33. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kumar, S.D.; Bang, J.K.; Shin, S.Y. Mechanisms of antimicrobial and antiendotoxin activities of a triazine-based amphipathic polymer. Biotechnol. Bioeng. 2020, 117, 3508–3521. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.; Barton, A.; Daher, K.A.; Harwig, S.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 1989, 84, 553–561. [Google Scholar] [PubMed]
- Basak, A.; Abouelhassan, Y.; Zuo, R.; Yousaf, H.; Ding, Y.; Huigens, R.W. Antimicrobial peptide-inspired NH125 analogues: Bacterial and fungal biofilm-eradicating agents and rapid killers of MRSA persisters. Org. Biomol. Chem. 2017, 15, 5503–5512. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.J.; Stremick, C.A.; Turner, R.J.; Allan, N.D.; Olson, M.E.; Ceri, H. Microtiter susceptibility testing of microbes growing on peg lids: A miniaturized biofilm model for high-throughput screening. Nat. Protoc. 2010, 5, 1236–1254. [Google Scholar] [CrossRef]
- Pandey, B.K.; Ahmad, A.; Asthana, N.; Azmi, S.; Srivastava, R.M.; Srivastava, S.; Verma, R.; Vishwakarma, A.L.; Ghosh, J.K. Cell-selective lysis by novel analogues of melittin against human red blood cells and Escherichia coli. Biochemistry 2010, 49, 7920–7929. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Pept. Sci. 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef]
- Gilbert, P.; Allison, D.; McBain, A. Biofilms in vitro andin vivo: Do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 2002, 92, 98S–110S. [Google Scholar] [CrossRef]

| Peptides | Sequences | Rt a (min) | Net Charge | Mass Analysis b | ||
|---|---|---|---|---|---|---|
| Mass (g/mol) | m/z Calculated | m/z Observed | ||||
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | 30.64 | 6 | 2847.49 | 949.83 | 949.8 |
| Mel-LX1 | GIGAVXKVLTTGLPALISWIKRKRQQ | 24.93 | 6 | 2847.49 | 949.83 | 949.8 |
| Mel-LX2 | GIGAVLKVXTTGLPALISWIKRKRQQ | 24.76 | 6 | 2847.49 | 949.83 | 949.7 |
| Mel-LX3 | GIGAVLKVLTTGXPALISWIKRKRQQ | 26.80 | 6 | 2847.49 | 949.83 | 949.7 |
| Mel-LX4 | GIGAVLKVLTTGLPAXISWIKRKRQQ | 24.75 | 6 | 2847.49 | 949.83 | 949.7 |
| Mel-LX5 | GIGAVXKVXTTGXPAXISWIKRKRQQ | 18.76 | 6 | 2847.49 | 949.83 | 949.7 |
| Microorganism | MIC (μM) | |||||
|---|---|---|---|---|---|---|
| Melittin | Mel-LX1 | Mel-LX2 | Mel-LX3 | Mel-LX4 | Mel-LX5 | |
| Gram-positive organisms | ||||||
| S. aureus (KCTC 1621) | 8 | 64 | 32 | 8 | 32 | >128 |
| S. epidermidis (KCTC 1917) | 8 | 32 | 32 | 16 | 32 | >64 |
| B. subtilis (KCTC 3068) | 16 | 16 | 16 | 16 | 32 | >64 |
| MRSA (CCARM 3090) | 4 | >128 | 128 | 4 | 128 | >128 |
| Gram-negative organisms | ||||||
| E. coli (KCTC 1682) | 8 | 64 | 32 | 16 | 32 | >128 |
| P. aeruginosa (KCTC 1637) | 16 | 64 | 64 | 16 | 64 | >64 |
| S. typhimurium (KCTC 1926) | 16 | 32 | 16 | 16 | 32 | >64 |
| MDRPA (CCARM 2095) | 8 | >128 | 128 | 8 | 128 | >128 |
| GM (μM) a | 10.5 | 98 | 56 | 12.5 | 60 | 192 |
| HC10 (μM) b | 2 | 64 | 128 | 64 | 128 | 128 |
| TI (HC10/GM) c | 0.19 | 0.65 | 2.28 | 5.12 | 2.13 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radhakrishnan, N.; Kumar, S.D.; Shin, S.-Y.; Yang, S. Enhancing Selective Antimicrobial and Antibiofilm Activities of Melittin through 6-Aminohexanoic Acid Substitution. Biomolecules 2024, 14, 699. https://doi.org/10.3390/biom14060699
Radhakrishnan N, Kumar SD, Shin S-Y, Yang S. Enhancing Selective Antimicrobial and Antibiofilm Activities of Melittin through 6-Aminohexanoic Acid Substitution. Biomolecules. 2024; 14(6):699. https://doi.org/10.3390/biom14060699
Chicago/Turabian StyleRadhakrishnan, Naveenkumar, Sukumar Dinesh Kumar, Song-Yub Shin, and Sungtae Yang. 2024. "Enhancing Selective Antimicrobial and Antibiofilm Activities of Melittin through 6-Aminohexanoic Acid Substitution" Biomolecules 14, no. 6: 699. https://doi.org/10.3390/biom14060699






