Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy and Study Selection
2.2. Data Extraction
2.3. Data Analysis
2.4. Quality Assessment
3. Results
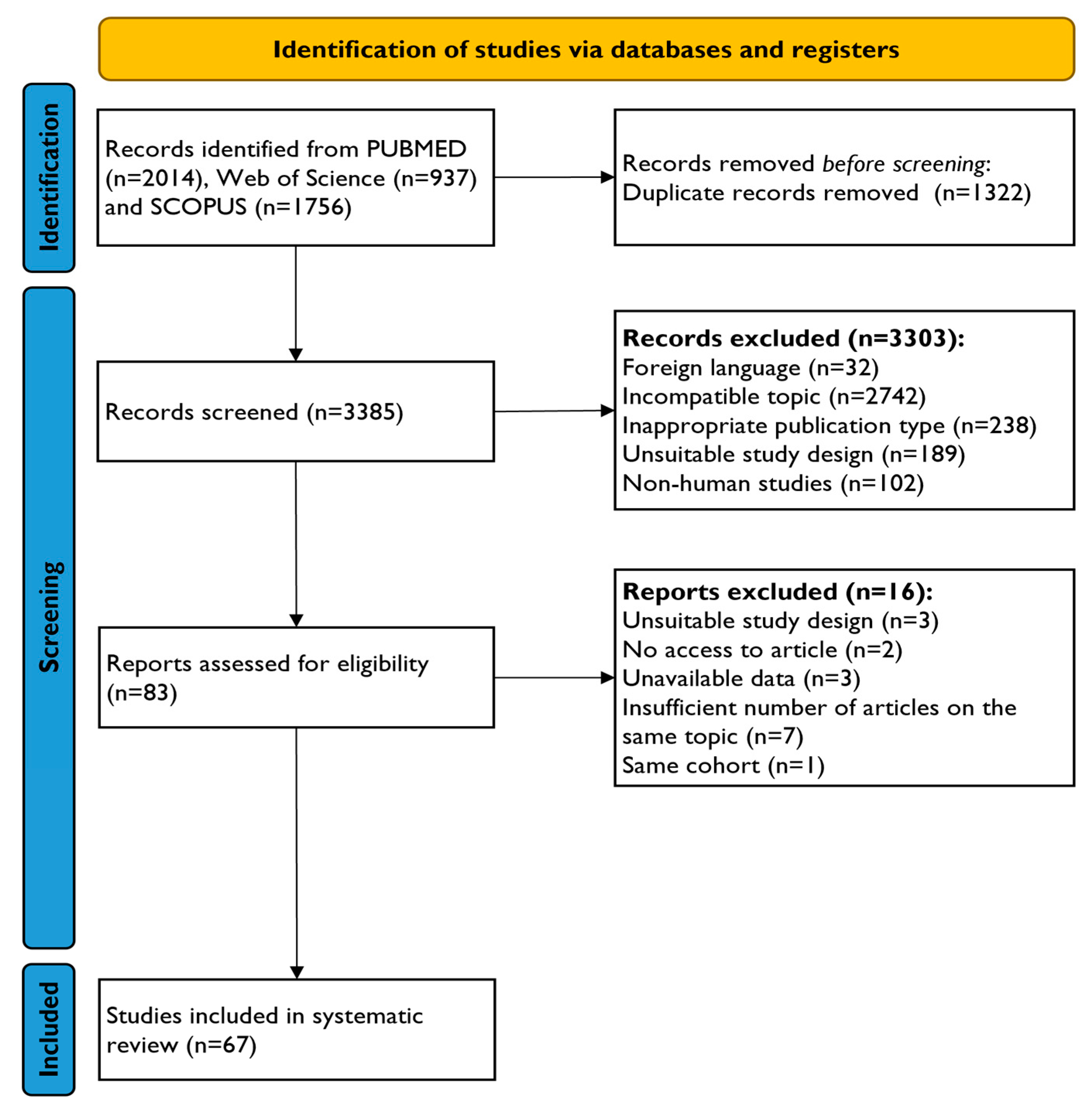
3.1. Study Selection and Characteristics
3.2. Methods of Metagenomic and Metabolomic Analysis
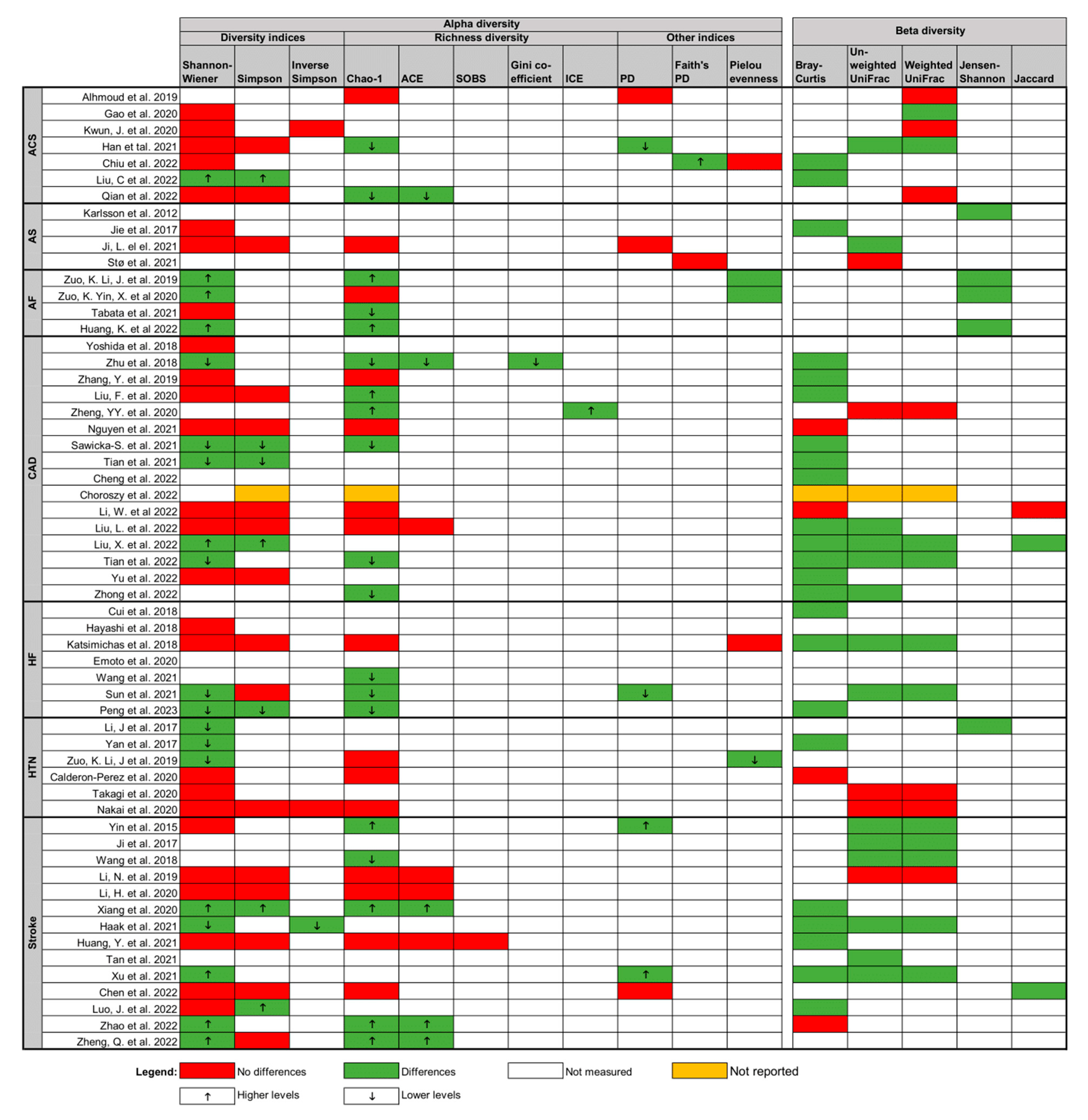
3.3. Qualitative Synthesis
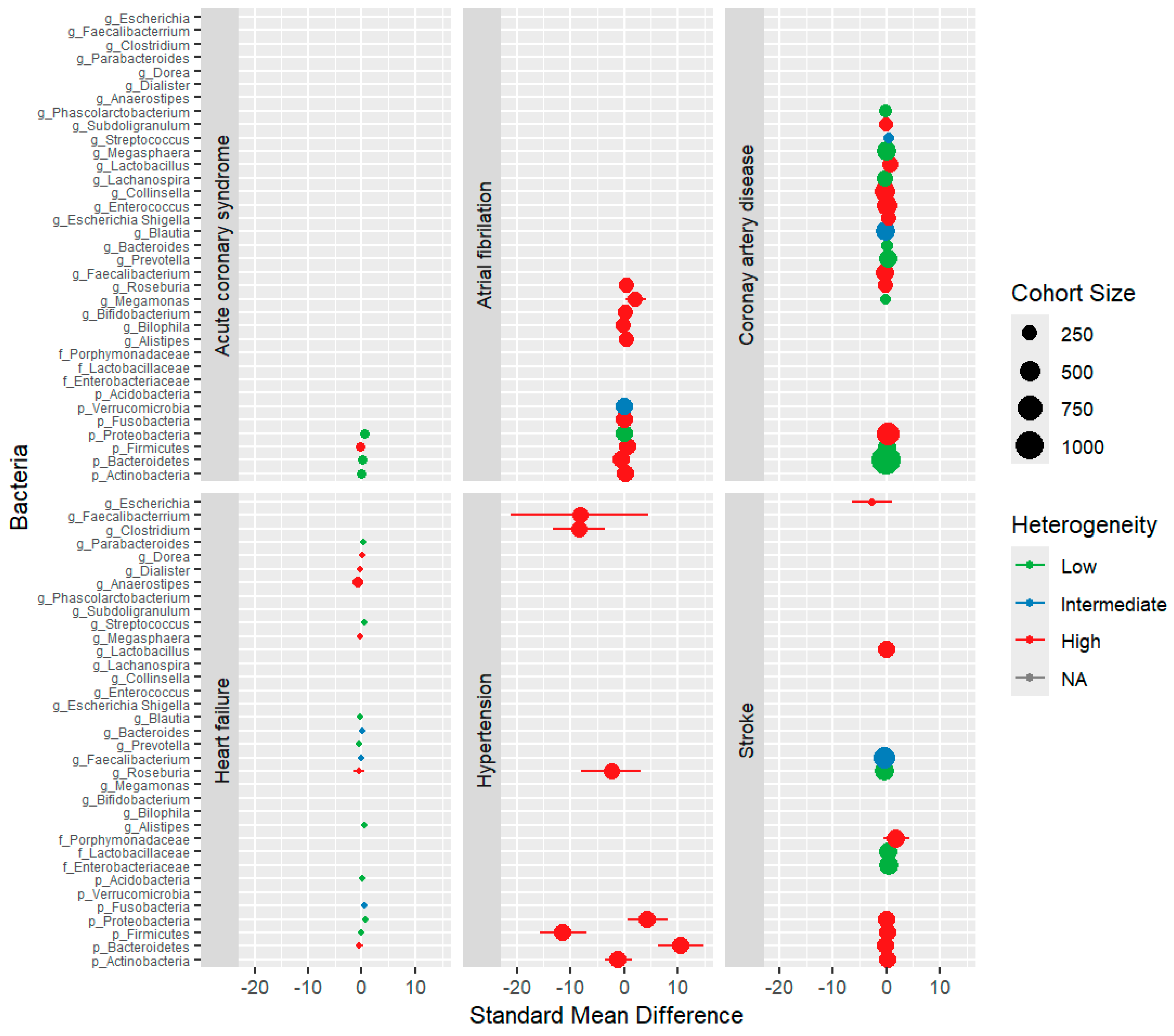
3.4. Quantitative Synthesis
4. Discussion
5. Strengths and Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Stilling, R.M.; Kennedy, P.J.; Stanton, C.; Cryan, J.F.; Dinan, T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014, 28, 1221–1238. [Google Scholar] [CrossRef]
- Yang, W.; Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 2021, 18, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T. Gut Microbiota Dysbiosis: Triggers, Consequences, Diagnostic and Therapeutic Options. Microorganisms 2022, 10, 578. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Kaur, G.; Kaur, A. Dysbiosis—An Etiological Factor for Cardiovascular Diseases and the Therapeutic Benefits of Gut Microflora. Adv. Gut Microbiome Res. 2023, 2023, 7451554. [Google Scholar] [CrossRef]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Violi, F.; Cammisotto, V.; Bartimoccia, S.; Pignatelli, P.; Carnevale, R.; Nocella, C. Gut-derived low-grade endotoxaemia, atherothrombosis and cardiovascular disease. Nat. Rev. Cardiol. 2023, 20, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Carresi, C.; Gliozzi, M.; Mollace, R.; Scarano, F.; Scicchitano, M.; Macrì, R.; Nucera, S.; Bosco, F.; Oppedisano, F.; et al. The Contribution of Gut Microbiota and Endothelial Dysfunction in the Development of Arterial Hypertension in Animal Models and in Humans. Int. J. Mol. Sci. 2022, 23, 3698. [Google Scholar] [CrossRef]
- Kazemian, N.; Mahmoudi, M.; Halperin, F.; Wu, J.C.; Pakpour, S. Gut microbiota and cardiovascular disease: Opportunities and challenges. Microbiome 2020, 8, 36. [Google Scholar] [CrossRef]
- Masenga, S.K.; Hamooya, B.; Hangoma, J.; Hayumbu, V.; Ertuglu, L.A.; Ishimwe, J.; Rahman, S.; Saleem, M.; Laffer, C.L.; Elijovich, F.; et al. Recent advances in modulation of cardiovascular diseases by the gut microbiota. J. Human. Hypertens. 2022, 36, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; Silva, C.; Ferreira, A.C.; Dourado, S.; Albuquerque, A.; Saraiva, F.; Batista, A.B.; Castro, P.; Leite-Moreira, P.; Barros, A.S.; et al. Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis. Available online: https://osf.io/49p75 (accessed on 25 May 2024).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, A. WebPlotDigitizer. Available online: https://automeris.io/WebPlotDigitizer (accessed on 31 May 2023).
- Wan, X.; Wang, W.; Liu, J.; Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 2014, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Cochrane. Review Manager Web (RevMan Web). Available online: https://revman.cochrane.org/ (accessed on 16 July 2023).
- National Heart, Lung, and Blood Institute. Study Quality Assessment Tools. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 26 June 2023).
- Alhmoud, T.; Kumar, A.; Lo, C.-C.; Al-Sadi, R.; Clegg, S.; Alomari, I.; Zmeili, T.; Gleasne, C.D.; McMurry, K.; Dichosa, A.E.K.; et al. Investigating intestinal permeability and gut microbiota roles in acute coronary syndrome patients. Human. Microbiome J. 2019, 13, 100059. [Google Scholar] [CrossRef] [PubMed]
- Büttner, P.; Okun, J.G.; Hauke, J.; Holzwirth, E.; Obradovic, D.; Hindricks, G.; Thiele, H.; Kornej, J. Trimethylamine N-oxide in atrial fibrillation progression. IJC Heart Vasc. 2020, 29, 100554. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Pérez, L.; Gosalbes, M.J.; Yuste, S.; Valls, R.M.; Pedret, A.; Llauradó, E.; Jimenez-Hernandez, N.; Artacho, A.; Pla-Pagà, L.; Companys, J.; et al. Gut metagenomic and short chain fatty acids signature in hypertension: A cross-sectional study. Sci. Rep. 2020, 10, 6436. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, S.; Zhang, Y.; Li, Y.; Zhang, X.; Ma, J.; Zou, X.; Yao, T.; Li, S.; Chen, J.; et al. Multi-omics reveals specific host metabolism-microbiome associations in intracerebral hemorrhage. Front. Cell. Infect. Microbiol. 2022, 12, 999627. [Google Scholar] [CrossRef]
- Cheng, Q.; Fan, C.; Liu, F.; Li, Y.; Hou, H.; Ma, Y.; Tan, Y.; Li, Y.; Hai, Y.; Wu, T.; et al. Structural and functional dysbiosis of gut microbiota in Tibetan subjects with coronary heart disease. Genomics 2022, 114, 110483. [Google Scholar] [CrossRef]
- Chiu, F.-C.; Tsai, C.-F.; Huang, P.-S.; Shih, C.-Y.; Tsai, M.-H.; Hwang, J.-J.; Wang, Y.-C.; Chuang, E.Y.; Tsai, C.-T.; Chang, S.-N. The Gut Microbiome, Seleno-Compounds, and Acute Myocardial Infarction. J. Clin. Med. 2022, 11, 1462. [Google Scholar] [CrossRef] [PubMed]
- Chong Nguyen, C.; Duboc, D.; Rainteau, D.; Sokol, H.; Humbert, L.; Seksik, P.; Bellino, A.; Abdoul, H.; Bouazza, N.; Treluyer, J.-M.; et al. Circulating bile acids concentration is predictive of coronary artery disease in human. Sci. Rep. 2021, 11, 22661. [Google Scholar] [CrossRef] [PubMed]
- Choroszy, M.; Sobieszczańska, B.; Litwinowicz, K.; Łaczmański, Ł.; Chmielarz, M.; Walczuk, U.; Roleder, T.; Radziejewska, J.; Wawrzyńska, M. Co-toxicity of Endotoxin and Indoxyl Sulfate, Gut-Derived Bacterial Metabolites, to Vascular Endothelial Cells in Coronary Arterial Disease Accompanied by Gut Dysbiosis. Nutrients 2022, 14, 424. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Ye, L.; Li, J.; Jin, L.; Wang, W.; Li, S.; Bao, M.; Wu, S.; Li, L.; Geng, B.; et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci. Rep. 2018, 8, 635. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Tian, Q.; Si, J.; Sun, Z.; Shali, S.; Xu, L.; Ren, D.; Chang, S.; Dong, X.; Zhao, H.; et al. Circulating metabolites from the choline pathway and acute coronary syndromes in a Chinese case-control study. Nutr. Metab. 2020, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zheng, S.; Shen, Z.; Luo, Y.; Hai, X. Trimethylamine N-Oxide is Associated with Heart Failure Risk in Patients with Preserved Ejection Fraction. Lab. Med. 2020, 52, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Emoto, T.; Hayashi, T.; Tabata, T.; Yamashita, T.; Watanabe, H.; Takahashi, T.; Gotoh, Y.; Kami, K.; Yoshida, N.; Saito, Y.; et al. Metagenomic analysis of gut microbiota reveals its role in trimethylamine metabolism in heart failure. Int. J. Cardiol. 2021, 338, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yan, K.-T.; Wang, J.-X.; Dou, J.; Wang, J.; Ren, M.; Ma, J.; Zhang, X.; Liu, Y. Gut microbial taxa as potential predictive biomarkers for acute coronary syndrome and post-STEMI cardiovascular events. Sci. Rep. 2020, 10, 2639. [Google Scholar] [CrossRef]
- Haak, B.W.; Westendorp, W.F.; van Engelen, T.S.R.; Brands, X.; Brouwer, M.C.; Vermeij, J.-D.; Hugenholtz, F.; Verhoeven, A.; Derks, R.J.; Giera, M.; et al. Disruptions of Anaerobic Gut Bacteria Are Associated with Stroke and Post-stroke Infection: A Prospective Case–Control Study. Transl. Stroke Res. 2021, 12, 581–592. [Google Scholar] [CrossRef]
- Han, Y.; Gong, Z.; Sun, G.; Xu, J.; Qi, C.; Sun, W.; Jiang, H.; Cao, P.; Ju, H. Dysbiosis of Gut Microbiota in Patients With Acute Myocardial Infarction. Front. Microbiol. 2021, 12, 680101. [Google Scholar] [CrossRef]
- Hayashi, T.; Yamashita, T.; Watanabe, H.; Kami, K.; Yoshida, N.; Tabata, T.; Emoto, T.; Sasaki, N.; Mizoguchi, T.; Irino, Y.; et al. Gut Microbiome and Plasma Microbiome-Related Metabolites in Patients With Decompensated and Compensated Heart Failure. Circ. J. 2018, 83, 182–192. [Google Scholar] [CrossRef]
- Huang, K.; Wang, Y.; Bai, Y.; Luo, Q.; Lin, X.; Yang, Q.; Wang, S.; Xin, H. Gut Microbiota and Metabolites in Atrial Fibrillation Patients and Their Changes after Catheter Ablation. Microbiol. Spectr. 2022, 10, e01077-21. [Google Scholar] [CrossRef]
- Huang, Y.; Shen, Z.; He, W. Identification of Gut Microbiome Signatures in Patients With Post-stroke Cognitive Impairment and Affective Disorder. Front. Aging Neurosci. 2021, 13, 706765. [Google Scholar] [CrossRef]
- Ji, L.; Chen, S.; Gu, G.; Zhou, J.; Wang, W.; Ren, J.; Wu, J.; Yang, D.; Zheng, Y. Exploration of Crucial Mediators for Carotid Atherosclerosis Pathogenesis Through Integration of Microbiome, Metabolome, and Transcriptome. Front. Physiol. 2021, 12, 645212. [Google Scholar] [CrossRef]
- Ji, W.; Zhu, Y.; Kan, P.; Cai, Y.; Wang, Z.; Wu, Z.; Yang, P. Analysis of intestinal microbial communities of cerebral infarction and ischemia patients based on high throughput sequencing technology and glucose and lipid metabolism. Mol. Med. Rep. 2017, 16, 5413–5417. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Katsimichas, T.; Ohtani, T.; Motooka, D.; Tsukamoto, Y.; Kioka, H.; Nakamoto, K.; Konishi, S.; Chimura, M.; Sengoku, K.; Miyawaki, H.; et al. Non-Ischemic Heart Failure With Reduced Ejection Fraction Is Associated With Altered Intestinal Microbiota. Circ. J. 2018, 82, 1640–1650. [Google Scholar] [CrossRef]
- Kwun, J.-S.; Kang, S.-H.; Lee, H.-J.; Park, H.-K.; Lee, W.-J.; Yoon, C.-H.; Suh, J.-W.; Cho, Y.-S.; Youn, T.-J.; Chae, I.-H. Comparison of thrombus, gut, and oral microbiomes in Korean patients with ST-elevation myocardial infarction: A case–control study. Exp. Mol. Med. 2020, 52, 2069–2079. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.; Pan, D.; Liu, Y.; Yan, X.; Tang, Y.; Tao, M.; Gong, L.; Zhang, T.; Woods, C.R.; et al. Dysbiosis characteristics of gut microbiota in cerebral infarction patients. Transl. Neurosci. 2020, 11, 124–133. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Sun, C.; Wu, X.; Lu, M.; Si, Y.; Ye, X.; Wang, T.; Yu, X.; Zhao, X.; et al. Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol. 2019, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, H.; Wang, S.; Han, K.; Liu, Y.; An, Z.; Wu, H.; Li, J.; Song, J.; Wu, W. Regional pattern and signatures of gut microbiota in rural residents with coronary heart disease: A metagenomic analysis. Front. Cell. Infect. Microbiol. 2022, 12, 1007161. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Sun, Z.; Shali, S.; Mei, Z.; Chang, S.; Mo, H.; Xu, L.; Pu, Y.; Guan, H.; Chen, G.C.; et al. The gut microbiome and microbial metabolites in acute myocardial infarction. J. Genet. Genom. 2022, 49, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Fan, C.; Zhang, L.; Li, Y.; Hou, H.; Ma, Y.; Fan, J.; Tan, Y.; Wu, T.; Jia, S.; et al. Alterations of Gut Microbiome in Tibetan Patients With Coronary Heart Disease. Front. Cell. Infect. Microbiol. 2020, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Luo, F. Alterations in the fecal microbiota and serum metabolome in unstable angina pectoris patients. Front. Biosci. (Landmark Ed.) 2022, 27, 100. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shen, M.; Yan, H.; Long, P.; Jiang, H.; Zhang, Y.; Zhou, L.; Yu, K.; Qiu, G.; Yang, H.; et al. Alternations in the gut microbiota and metabolome with newly diagnosed unstable angina. J. Genet. Genom. 2022, 49, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, Y.; Tang, G.; Li, Z.; Yang, X.; Shang, X.; Huang, T.; Huang, G.; Wang, L.; Han, Y.; et al. Gut microbiota composition reflects disease progression, severity and outcome, and dysfunctional immune responses in patients with hypertensive intracerebral hemorrhage. Front. Immunol. 2022, 13, 869846. [Google Scholar] [CrossRef]
- Nakai, M.; Ribeiro, R.V.; Stevens, B.R.; Gill, P.; Muralitharan, R.R.; Yiallourou, S.; Muir, J.; Carrington, M.; Head, G.A.; Kaye, D.M.; et al. Essential Hypertension Is Associated with Changes in Gut Microbial Metabolic Pathways: A Multisite Analysis of Ambulatory Blood Pressure. Hypertension 2021, 78, 804–815. [Google Scholar] [CrossRef]
- Nie, J.; Xie, L.; Zhao, B.-x.; Li, Y.; Qiu, B.; Zhu, F.; Li, G.-f.; He, M.; Wang, Y.; Wang, B.; et al. Serum Trimethylamine N-Oxide Concentration Is Positively Associated With First Stroke in Hypertensive Patients. Stroke 2018, 49, 2021–2028. [Google Scholar] [CrossRef]
- Peng, J.; Gong, H.; Lyu, X.; Liu, Y.; Li, S.; Tan, S.; Dong, L.; Zhang, X. Characteristics of the fecal microbiome and metabolome in older patients with heart failure and sarcopenia. Front. Cell. Infect. Microbiol. 2023, 13, 1127041. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Liu, A.; Liang, C.; He, L.; Xu, Z.; Tang, S. Analysis of gut microbiota in patients with acute myocardial infarction by 16S rRNA sequencing. Ann. Transl. Med. 2022, 10, 1340. [Google Scholar] [CrossRef] [PubMed]
- Sawicka-Smiarowska, E.; Bondarczuk, K.; Bauer, W.; Niemira, M.; Szalkowska, A.; Raczkowska, J.; Kwasniewski, M.; Tarasiuk, E.; Dubatowka, M.; Lapinska, M.; et al. Gut Microbiome in Chronic Coronary Syndrome Patients. J. Clin. Med. 2021, 10, 5074. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Okun, J.G.; Schwarz, K.V.; Hauke, J.; Zorn, M.; Nürnberg, C.; Ungerer, M.; Ringleb, P.A.; Mundiyanapurath, S. Trimethylamine-N-oxide is elevated in the acute phase after ischaemic stroke and decreases within the first days. Eur. J. Neurol. 2020, 27, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Stø, K.; Valeur, J.; Ueland, T.; Malmstrøm, G.H.; Bjerkeli, V.; Trøseid, M.; Hov, J.R.; Holm, K.; Vestad, B.; Halvorsen, B.; et al. Fecal level of butyric acid, a microbiome-derived metabolite, is increased in patients with severe carotid atherosclerosis. Sci. Rep. 2022, 12, 22378. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Y.; Yin, J.; Peng, X.; Zhou, L.; Huang, S.; Wen, Y.; Cao, B.; Chen, L.; Li, X.; et al. Association of Gut Microbiota-Dependent Metabolite Trimethylamine N-Oxide with First Ischemic Stroke. J. Atheroscler. Thromb. 2021, 28, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Du, D.; Fu, T.; Han, Y.; Li, P.; Ju, H. Alterations of the Gut Microbiota in Patients With Severe Chronic Heart Failure. Front. Microbiol. 2022, 12, 813289. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Yamashita, T.; Hosomi, K.; Park, J.; Hayashi, T.; Yoshida, N.; Saito, Y.; Fukuzawa, K.; Konishi, K.; Murakami, H.; et al. Gut microbial composition in patients with atrial fibrillation: Effects of diet and drugs. Heart Vessel. 2021, 36, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Naito, Y.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Kamada, K.; Ishikawa, T.; Inoue, R.; Okuda, K.; Tsujimoto, Y.; et al. Changes in the Gut Microbiota are Associated with Hypertension, Hyperlipidemia, and Type 2 Diabetes Mellitus in Japanese Subjects. Nutrients 2020, 12, 2996. [Google Scholar] [CrossRef]
- Tan, C.; Wu, Q.; Wang, H.; Gao, X.; Xu, R.; Cui, Z.; Zhu, J.; Zeng, X.; Zhou, H.; He, Y.; et al. Dysbiosis of Gut Microbiota and Short-Chain Fatty Acids in Acute Ischemic Stroke and the Subsequent Risk for Poor Functional Outcomes. JPEN J. Parenter. Enter. Nutr. 2021, 45, 518–529. [Google Scholar] [CrossRef]
- Tang, Y.; Zou, Y.; Cui, J.; Ma, X.; Zhang, L.; Yu, S.; Qiu, L. Analysis of two intestinal bacterial metabolites (trimethylamine N-oxide and phenylacetylglutamine) in human serum samples of patients with T2DM and AMI using a liquid chromatography tandem mass spectrometry method. Clin. Chim. Acta 2022, 536, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Liu, H.; Feng, S.; Wang, H.; Wang, Y.; Wang, Y.; Liang, L.; Xu, H.; Xing, H.; Zhang, S. Gut microbiota dysbiosis in stable coronary artery disease combined with type 2 diabetes mellitus influences cardiovascular prognosis. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 1454–1466. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Liu, H.-H.; Feng, S.-Q.; Wang, Y.-F.; Wang, Y.-Y.; Chen, Y.-X.; Wang, H.; Zhang, S.-Y. Gut microbiota metabolic characteristics in coronary artery disease patients with hyperhomocysteine. J. Microbiol. 2022, 60, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Trøseid, M.; Ueland, T.; Hov, J.R.; Svardal, A.; Gregersen, I.; Dahl, C.P.; Aakhus, S.; Gude, E.; Bjørndal, B.; Halvorsen, B.; et al. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J. Intern. Med. 2015, 277, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, X.; Yao, X.; Cheng, X.; Zhu, Y. The characteristics analysis of intestinal microecology on cerebral infarction patients and its correlation with apolipoprotein E. Medicine 2018, 97, e12805. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, Z.; Ferrari, M.W.; Liu, Y.; Li, C.; Zhang, T.; Lyu, G. The Correlation between Gut Microbiota and Serum Metabolomic in Elderly Patients with Chronic Heart Failure. Mediat. Inflamm. 2021, 2021, 5587428. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Lou, Y.; Liu, L.; Liu, Y.; Zhang, W.; Deng, J.; Guan, Y.; She, M.; You, X.; Liu, M.; et al. Gut Microbiotic Features Aiding the Diagnosis of Acute Ischemic Stroke. Front. Cell. Infect. Microbiol. 2020, 10, 587284. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-J.; Wang, K.-C.; Yuan, L.-B.; Li, H.-F.; Xu, Y.-Y.; Wei, L.-Y.; Chen, L.; Jin, K.-K.; Lin, Q.-Q. Compositional and functional alterations of gut microbiota in patients with stroke. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 3434–3448. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Gu, Y.; Li, X.; Yang, W.; Jia, L.; Chen, C.; Han, X.; Huang, Y.; Zhao, L.; Li, P.; et al. Alterations of the Gut Microbiome in Hypertension. Front. Cell. Infect. Microbiol. 2017, 7, 381. [Google Scholar] [CrossRef]
- Yin, J.; Liao, S.X.; He, Y.; Wang, S.; Xia, G.H.; Liu, F.T.; Zhu, J.J.; You, C.; Chen, Q.; Zhou, L.; et al. Dysbiosis of Gut Microbiota with Reduced Trimethylamine-N-Oxide Level in Patients with Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 2015, 4, e002699. [Google Scholar] [CrossRef]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides vulgatus and Bacteroides dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, L.; Deng, Y.; Zhang, G.; Jiang, M.; Huang, H.; Li, C.; Lv, Z.; Zhou, Y.; Liu, X. The relationship between the number of stenotic coronary arteries and the gut microbiome in coronary heart disease patients. Front. Cell. Infect. Microbiol. 2022, 12, 903828. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Cai, J.; Lei, A.; Liu, C.; Lin, R.; Jia, L.; Fu, Y. Gut microbial metabolite TMAO portends prognosis in acute ischemic stroke. J. Neuroimmunol. 2021, 354, 577526. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Wang, X.; Ren, X.; Liu, Y. Changes of intestinal bacterial microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. BMC Genom. 2019, 20, 862. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, C.; Peng, S.; Zhu, X.; Zhang, Z.; Zhao, Y.; Zhang, J.; Zhao, G.; Zhang, T.; Heng, X.; et al. Pivotal interplays between fecal metabolome and gut microbiome reveal functional signatures in cerebral ischemic stroke. J. Transl. Med. 2022, 20, 459. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Chen, Y.; Zhai, Y.; Meng, L.; Liu, H.; Tian, H.; Feng, R.; Wang, J.; Zhang, R.; Sun, K.; et al. Gut Dysbiosis Is Associated With the Severity of Cryptogenic Stroke and Enhanced Systemic Inflammatory Response. Front. Immunol. 2022, 13, 836820. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.Y.; Wu, T.T.; Liu, Z.Q.; Li, A.; Guo, Q.Q.; Ma, Y.Y.; Zhang, Z.L.; Xun, Y.L.; Zhang, J.C.; Wang, W.R.; et al. Gut Microbiome-Based Diagnostic Model to Predict Coronary Artery Disease. J. Agric. Food Chem. 2020, 68, 3548–3557. [Google Scholar] [CrossRef]
- Zhong, J.; Wu, D.; Zeng, Y.; Wu, G.; Zheng, N.; Huang, W.; Li, Y.; Tao, X.; Zhu, W.; Sheng, L.; et al. The Microbial and Metabolic Signatures of Patients with Stable Coronary Artery Disease. Microbiol. Spectr. 2022, 10, e02467-22. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gao, R.; Zhang, Y.; Pan, D.; Zhu, Y.; Zhang, X.; Yang, R.; Jiang, R.; Xu, Y.; Qin, H. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol. Genom. 2018, 50, 893–903. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Li, K.; Hu, C.; Gao, Y.; Chen, M.; Hu, R.; Liu, Y.; Chi, H.; Wang, H.; et al. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. GigaScience 2019, 8, giz058. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Xu, Q.; Hu, C.; Gao, Y.; Chen, M.; Hu, R.; Liu, Y.; Chi, H.; Yin, Q.; et al. Dysbiotic gut microbes may contribute to hypertension by limiting vitamin D production. Clin. Cardiol. 2019, 42, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Zuo, K.; Yin, X.; Li, K.; Zhang, J.; Wang, P.; Jiao, J.; Liu, Z.; Liu, X.; Liu, J.; Li, J.; et al. Different Types of Atrial Fibrillation Share Patterns of Gut Microbiota Dysbiosis. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Liu, H.; Pan, L.-L.; Lv, S.; Yang, Q.; Zhang, H.; Chen, W.; Lv, Z.; Sun, J. Alterations of Gut Microbiota and Blood Lipidome in Gestational Diabetes Mellitus With Hyperlipidemia. Front. Physiol. 2019, 10, 1015. [Google Scholar] [CrossRef]
- Xu, D.-J.; Wang, K.; Yuan, L.-B.; Lin, Q.-Q.; Li, H.-F.; Xu, Y.-Y.; Wei, L.-Y.; Chen, L.; Jin, K.-K. Dysbiosis of Gut Microbiota with Increased Trimethylamine N-oxide Level in Patients with Large Artery Atherosclerotic and Cardioembolic Strokes. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Yoshida, N.; Yamashita, T.; Hirata, K.-i. Gut Microbiome and Cardiovascular Diseases. Diseases 2018, 6, 56. [Google Scholar] [CrossRef]
- Zuo, K.; Li, J.; Wang, P.; Liu, Y.; Liu, Z.; Yin, X.; Liu, X.; Yang, X. Duration of Persistent Atrial Fibrillation Is Associated with Alterations in Human Gut Microbiota and Metabolic Phenotypes. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Trøseid, M.; Andersen, G.; Broch, K.; Hov, J.R. The gut microbiome in coronary artery disease and heart failure: Current knowledge and future directions. EBioMedicine 2020, 52, 102649. [Google Scholar] [CrossRef]
- Khan, I.; Khan, I.; Jianye, Z.; Xiaohua, Z.; Khan, M.; Hilal, M.G.; Kakakhel, M.A.; Mehmood, A.; Lizhe, A.; Zhiqiang, L. Exploring blood microbial communities and their influence on human cardiovascular disease. J. Clin. Lab. Anal. 2022, 36, e24354. [Google Scholar] [CrossRef]
- Sayols-Baixeras, S.; Dekkers, K.F.; Hammar, U.; Baldanzi, G.; Lin, Y.-T.; Ahmad, S.; Nguyen, D.; Varotsis, G.; Pita, S.; Nielsen, N.; et al. Streptococcus species abundance in the gut is linked to subclinical coronary atherosclerosis in 8,973 participants from the SCAPIS cohort. medRxiv 2022. [Google Scholar] [CrossRef]
- Ott, S.J.; El Mokhtari, N.E.; Musfeldt, M.; Hellmig, S.; Freitag, S.; Rehman, A.; Kühbacher, T.; Nikolaus, S.; Namsolleck, P.; Blaut, M.; et al. Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation 2006, 113, 929–937. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Lui, G.K.; Fernandes, S.; McElhinney, D.B. Management of cardiovascular risk factors in adults with congenital heart disease. J. Am. Heart Assoc. 2014, 3, e001076. [Google Scholar] [CrossRef] [PubMed]
- Janeiro, M.H.; Ramírez, M.J.; Milagro, F.I.; Martínez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, 1398. [Google Scholar] [CrossRef]
- Li, Q.; Larouche-Lebel, É.; Loughran, K.A.; Huh, T.P.; Suchodolski, J.S.; Oyama, M.A. Gut Dysbiosis and Its Associations with Gut Microbiota-Derived Metabolites in Dogs with Myxomatous Mitral Valve Disease. mSystems 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Veys, K.; Fan, Z.; Ghobrial, M.; Bouché, A.; García-Caballero, M.; Vriens, K.; Conchinha, N.; Seuwen, A.; Schlegel, F.; Gorski, T.; et al. Role of the GLUT1 Glucose Transporter in Postnatal CNS Angiogenesis and Blood-Brain Barrier Integrity. Circ. Res. 2020, 127, 466–482. [Google Scholar] [CrossRef]
- Biruete, A.; Allen, J.M.; Kistler, B.M.; Jeong, J.H.; Fitschen, P.J.; Swanson, K.S.; Wilund, K.R. Gut Microbiota and Cardiometabolic Risk Factors in Hemodialysis Patients: A Pilot Study. Top. Clin. Nutr. 2019, 34, 153–160. [Google Scholar] [CrossRef]
- van den Munckhof, I.C.L.; Kurilshikov, A.; ter Horst, R.; Riksen, N.P.; Joosten, L.A.B.; Zhernakova, A.; Fu, J.; Keating, S.T.; Netea, M.G.; de Graaf, J.; et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: A systematic review of human studies. Obes. Rev. 2018, 19, 1719–1734. [Google Scholar] [CrossRef]
- Jie, Z.; Zhu, Q.; Zou, Y.; Wu, Q.; Qin, M.; He, D.; Lin, X.; Tong, X.; Zhang, J.; Jie, Z.; et al. A consortium of three-bacteria isolated from human feces inhibits formation of atherosclerotic deposits and lowers lipid levels in a mouse model. iScience 2023, 26, 106960. [Google Scholar] [CrossRef] [PubMed]
- Brahe, L.K.; Le Chatelier, E.; Prifti, E.; Pons, N.; Kennedy, S.; Hansen, T.; Pedersen, O.; Astrup, A.; Ehrlich, S.D.; Larsen, L.H. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes 2015, 5, e159. [Google Scholar] [CrossRef]
- Huang, X.; Fang, S.; Yang, H.; Gao, J.; He, M.; Ke, S.; Zhao, Y.; Chen, C.; Huang, L. Evaluating the contribution of gut microbiome to the variance of porcine serum glucose and lipid concentration. Sci. Rep. 2017, 7, 14928. [Google Scholar] [CrossRef]
- Canyelles, M.; Borràs, C.; Rotllan, N.; Tondo, M.; Escolà-Gil, J.C.; Blanco-Vaca, F. Gut Microbiota-Derived TMAO: A Causal Factor Promoting Atherosclerotic Cardiovascular Disease? Int. J. Mol. Sci. 2023, 24, 1940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-Q.; Wang, Y.-J.; Zhang, A.; Ding, Y.-J.; Zhang, X.-N.; Jia, Q.-J.; Zhu, Y.-P.; Li, Y.-Y.; Lv, S.-C.; Zhang, J.-P. TMA/TMAO in Hypertension: Novel Horizons and Potential Therapies. J. Cardiovasc. Transl. Res. 2021, 14, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Jiménez-Pranteda, M.L.; Chilloux, J.; Brial, F.; Myridakis, A.; Aranias, T.; Magnan, C.; Gibson, G.R.; Sanderson, J.D.; Nicholson, J.K.; et al. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. Microbiome 2018, 6, 73. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, D.; Silva, C.; Ferreira, A.C.; Dourado, S.; Albuquerque, A.; Saraiva, F.; Batista, A.B.; Castro, P.; Leite-Moreira, A.; Barros, A.S.; et al. Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis. Biomolecules 2024, 14, 731. https://doi.org/10.3390/biom14060731
Martins D, Silva C, Ferreira AC, Dourado S, Albuquerque A, Saraiva F, Batista AB, Castro P, Leite-Moreira A, Barros AS, et al. Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis. Biomolecules. 2024; 14(6):731. https://doi.org/10.3390/biom14060731
Chicago/Turabian StyleMartins, Diana, Cláudia Silva, António Carlos Ferreira, Sara Dourado, Ana Albuquerque, Francisca Saraiva, Ana Beatriz Batista, Pedro Castro, Adelino Leite-Moreira, António S. Barros, and et al. 2024. "Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis" Biomolecules 14, no. 6: 731. https://doi.org/10.3390/biom14060731
APA StyleMartins, D., Silva, C., Ferreira, A. C., Dourado, S., Albuquerque, A., Saraiva, F., Batista, A. B., Castro, P., Leite-Moreira, A., Barros, A. S., & Miranda, I. M. (2024). Unravelling the Gut Microbiome Role in Cardiovascular Disease: A Systematic Review and a Meta-Analysis. Biomolecules, 14(6), 731. https://doi.org/10.3390/biom14060731







