An Early Gestation Plasma Inflammasome in Rural Bangladeshi Women
Abstract
:1. Introduction
2. Materials and Methods
2.1. JiVitA-3 Trial
2.2. Biochemical Substudy
2.3. Proteomics Substudy
2.4. Statistical Analysis
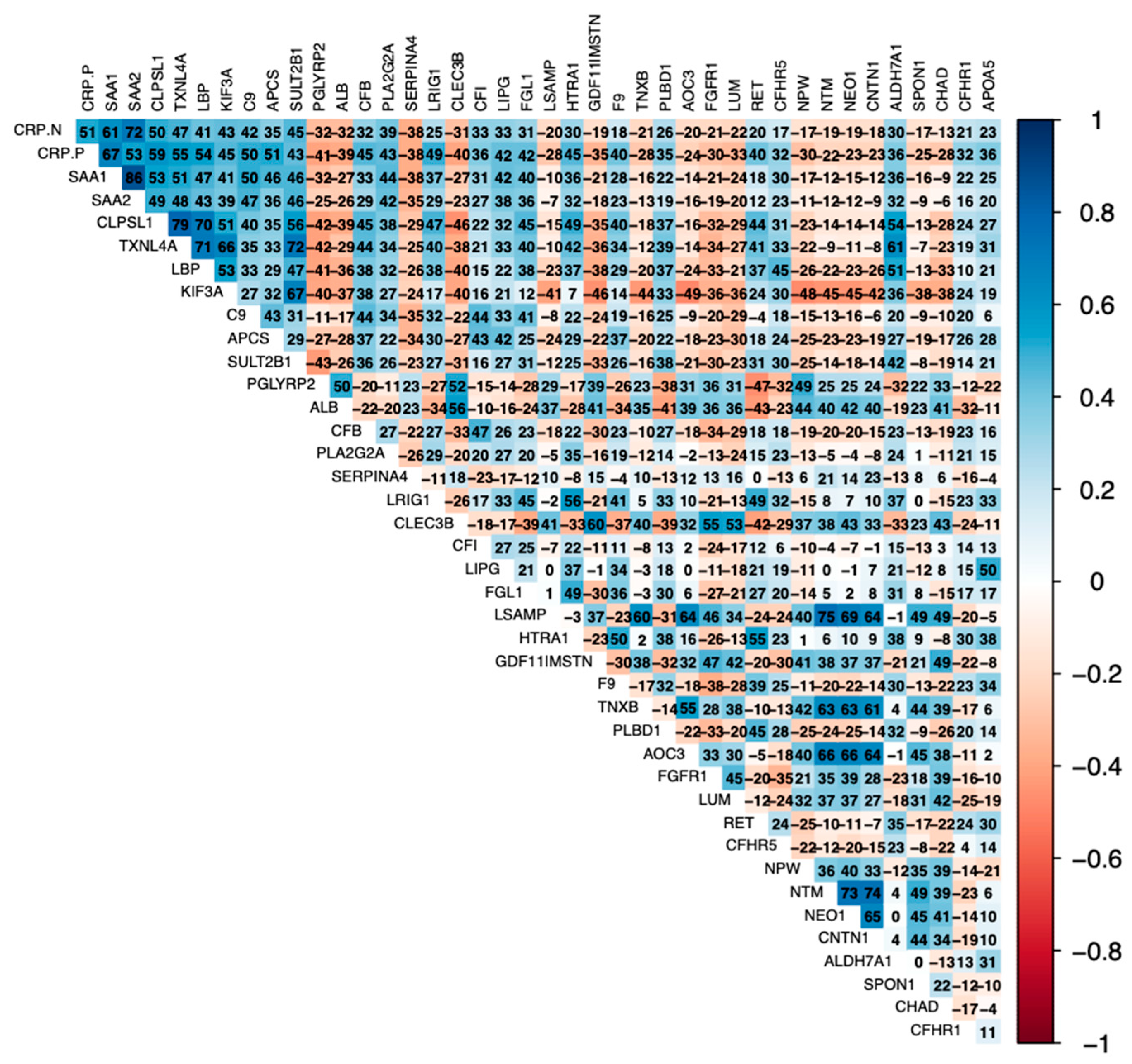
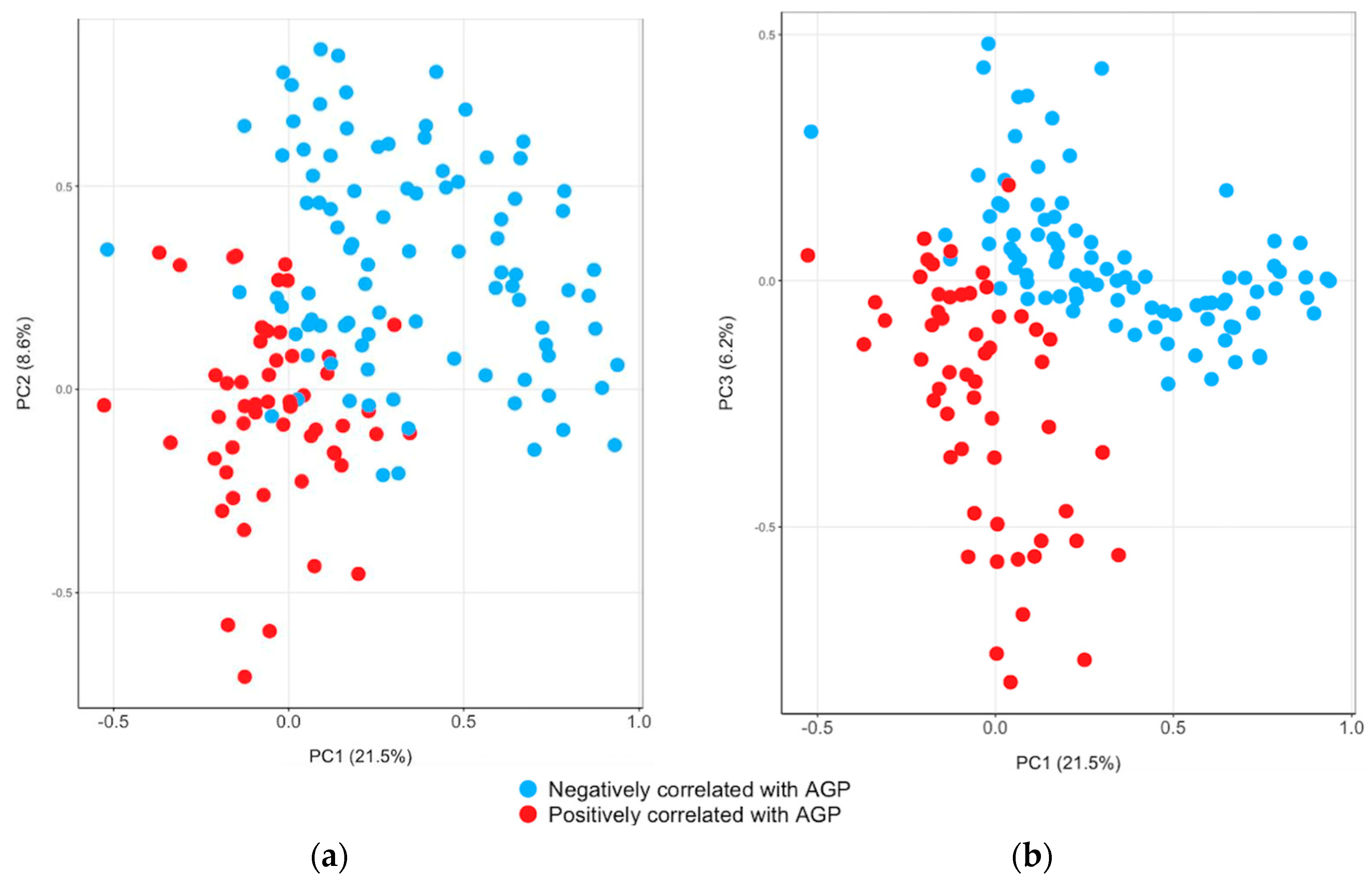
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mor, G.; Cardenas, I.; Abrahams, V.; Guller, S. Inflammation and Pregnancy: The Role of the Immune System at the Implantation Site. Ann. N. Y. Acad. Sci. 2011, 1221, 80. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Jauniaux, E.; Charnock-Jones, D.S. The Influence of the Intrauterine Environment on Human Placental Development. Int. J. Dev. Biol. 2010, 54, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Muter, J.; Lynch, V.J.; McCoy, R.C.; Brosens, J.J. Human Embryo Implantation. Development 2023, 150, dev201507. [Google Scholar] [CrossRef] [PubMed]
- Förger, F.; Villiger, P.M. Immunological Adaptations in Pregnancy That Modulate Rheumatoid Arthritis Disease Activity. Nat. Rev. Rheumatol. 2020, 16, 113–122. [Google Scholar] [CrossRef] [PubMed]
- de Rivero Vaccari, J.P. The Inflammasome in Reproductive Biology: A Promising Target for Novel Therapies. Front. Endocrinol. 2020, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Baumann, H.; Gauldie, J. The Acute Phase Response. Immunol. Today 1994, 15, 74–80. [Google Scholar] [CrossRef]
- Sauder, M.W.; Lee, S.E.; Schulze, K.J.; Christian, P.; Wu, L.S.F.; Khatry, S.K.; LeClerq, S.C.; Adhikari, R.K.; Groopman, J.D.; West, K.P. Inflammation throughout Pregnancy and Fetal Growth Restriction in Rural Nepal. Epidemiol. Infect. 2019, 147, e258. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Suchdev, P.S.; Krebs, N.F.; Hess, S.Y.; Wessells, K.R.; Ismaily, S.; Rahman, S.; Wieringa, F.T.; Williams, A.M.; Brown, K.H.; et al. Adjusting Plasma or Serum Zinc Concentrations for Inflammation: Biomarkers Reflecting Inflammation and Nutritional Determinants of Anemia (BRINDA) Project. Am. J. Clin. Nutr. 2020, 111, 927–937. [Google Scholar] [CrossRef]
- Ghosh, S.; Kurpad, A.V.; Sachdev, H.S.; Thomas, T. Inflammation Correction in Micronutrient Deficiency with Censored Inflammatory Biomarkers. Am. J. Clin. Nutr. 2021, 113, 47–54. [Google Scholar] [CrossRef]
- Namaste, S.M.; Aaron, G.J.; Varadhan, R.; Peerson, J.M.; Suchdev, P.S. Methodologic Approach for the Biomarkers Reflecting Inflammation and Nutritional Determinants of Anemia (BRINDA) Project. Am. J. Clin. Nutr. 2017, 106, 333S–347S. [Google Scholar] [CrossRef]
- Thurnham, D.I.; McCabe, G.P.; Northrop-Clewes, C.A.; Nestel, P. Effects of Subclinical Infection on Plasma Retinol Concentrations and Assessment of Prevalence of Vitamin A Deficiency: Meta-Analysis. Lancet 2003, 362, 2052–2058. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Innate Immunity. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Chaplin, D.D. Overview of the Immune Response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Singh-Manoux, A.; Shipley, M.J.; Bell, J.A.; Canonico, M.; Elbaz, A.; Kivimäki, M. Association between Inflammatory Biomarkers and All-Cause, Cardiovascular and Cancer-Related Mortality. CMAJ 2017, 189, E384–E390. [Google Scholar] [CrossRef] [PubMed]
- Lauer, J.M.; Ghosh, S.; Ausman, L.M.; Webb, P.; Bashaasha, B.; Agaba, E.; Turyashemererwa, F.M.; Tran, H.Q.; Gewirtz, A.T.; Erhardt, J.; et al. Markers of Environmental Enteric Dysfunction Are Associated with Poor Growth and Iron Status in Rural Ugandan Infants. J. Nutr. 2020, 150, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Raiten, D.J.; Ashour, F.A.S.; Ross, A.C.; Meydani, S.N.; Dawson, H.D.; Stephensen, C.B.; Brabin, B.J.; Suchdev, P.S.; van Ommen, B. Inflammation and Nutritional Science for Programs/Policies and Interpretation of Research Evidence (INSPIRE). J. Nutr. 2015, 145, 1039S–1108S. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Kivimaki, M.; Langenberg, C.; Hingorani, A.D.; Casas, J.P.; Bouchard, C.; Jonasson, C.; Sarzynski, M.A.; Shipley, M.J.; Alexander, L.; et al. Plasma Protein Patterns as Comprehensive Indicators of Health. Nat. Med. 2019, 25, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Ganz, P.; Heidecker, B.; Hveem, K.; Jonasson, C.; Kato, S.; Segal, M.R.; Sterling, D.G.; Williams, S.A. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes among Patients with Stable Coronary Heart Disease. JAMA 2016, 315, 2532–2541. [Google Scholar] [CrossRef]
- Lee, S.E.; Schulze, K.; West, K.P. Rainer Gross Award Lecture 2018: The Childhood Plasma Proteome: Discovering Its Applications in Public Health Nutrition. Food Nutr. Bull. 2019, 40, 144–150. [Google Scholar] [CrossRef]
- Lee, S.E.; West, K.P.W., Jr.; Cole, R.N.; Schulze, K.J.; Christian, P.; Wu, L.S.-F.; Yager, J.D.; Groopman, J.; Ruczinski, I. Plasma Proteome Biomarkers of Inflammation in School Aged Children in Nepal. PLoS ONE 2015, 10, e0144279. [Google Scholar] [CrossRef]
- Scholl, P.F.; Cole, R.N.; Ruczinski, I.; Gucek, M.; Diez, R.; Rennie, A.; Nathasingh, C.; Schulze, K.; Christian, P.; Yager, J.D.; et al. Maternal Serum Proteome Changes between the First and Third Trimester of Pregnancy in Rural Southern Nepal. Placenta 2012, 33, 424–432. [Google Scholar] [CrossRef]
- Tarca, A.L.; Romero, R.; Bhatti, G.; Gotsch, F.; Done, B.; Gudicha, D.W.; Gallo, D.M.; Jung, E.; Pique-Regi, R.; Berry, S.M.; et al. Human Plasma Proteome during Normal Pregnancy. J. Proteome Res. 2022, 21, 2687–2702. [Google Scholar] [CrossRef]
- West, K.P.; Christian, P.; Labrique, A.B.; Rashid, M.; Shamim, A.A.; Klemm, R.D.W.; Massie, A.B.; Mehra, S.; Schulze, K.J.; Ali, H.; et al. Effects of Vitamin A or Beta Carotene Supplementation on Pregnancy-Related Mortality and Infant Mortality in Rural Bangladesh: A Cluster Randomized Trial. JAMA 2011, 305, 1986–1995. [Google Scholar] [CrossRef]
- Labrique, A.B.; Christian, P.; Klemm, R.D.W.; Rashid, M.; Shamim, A.A.; Massie, A.; Schulze, K.; Hackman, A.; West, K.P. A Cluster-Randomized, Placebo-Controlled, Maternal Vitamin A or Beta-Carotene Supplementation Trial in Bangladesh: Design and Methods. Trials 2011, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Gunnsteinsson, S.; Labrique, A.B.; West, K.P.; Christian, P.; Mehra, S.; Shamim, A.A.; Rashid, M.; Katz, J.; Klemm, R.D.W. Constructing Indices of Rural Living Standards in Northwestern Bangladesh. J. Health Popul. Nutr. 2010, 28, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.J.; Gernand, A.D.; Khan, A.Z.; Wu, L.S.-F.; Mehra, S.; Shaikh, S.; Ali, H.; Shamim, A.A.; Sungpuag, P.; Udomkesmalee, E.; et al. Newborn Micronutrient Status Biomarkers in a Cluster-Randomized Trial of Antenatal Multiple Micronutrient Compared with Iron Folic Acid Supplementation in Rural Bangladesh. Am. J. Clin. Nutr. 2020, 112, 1328–1337. [Google Scholar] [CrossRef]
- Gold, L.; Walker, J.J.; Wilcox, S.K.; Williams, S. Advances in Human Proteomics at High Scale with the SOMAscan Proteomics Platform. New Biotechnol. 2012, 29, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Bartalena, L. Thyroid Hormone-Binding Proteins. In Encyclopedia of Endocrine Diseases; Martini, L., Ed.; Elsevier: New York, NY, USA, 2004; pp. 474–479. ISBN 978-0-12-475570-3. [Google Scholar]
- Selby, C. Sex Hormone Binding Globulin: Origin, Function and Clinical Significance. Ann. Clin. Biochem. 1990, 27 Pt 6, 532–541. [Google Scholar] [CrossRef]
- Rosales, F.J.; Ritter, S.J.; Zolfaghari, R.; Smith, J.E.; Ross, A.C. Effects of Acute Inflammation on Plasma Retinol, Retinol-Binding Protein, and Its mRNA in the Liver and Kidneys of Vitamin A-Sufficient Rats. J. Lipid Res. 1996, 37, 962–971. [Google Scholar] [CrossRef]
- Khovidhunkit, W.; Kim, M.-S.; Memon, R.A.; Shigenaga, J.K.; Moser, A.H.; Feingold, K.R.; Grunfeld, C. Effects of Infection and Inflammation on Lipid and Lipoprotein Metabolism: Mechanisms and Consequences to the Host. J. Lipid Res. 2004, 45, 1169–1196. [Google Scholar] [CrossRef]
- Liao, C.-H.; Li, H.-Y.; Yu, H.-J.; Chiang, H.-S.; Lin, M.-S.; Hua, C.-H.; Ma, W.-Y. Low Serum Sex Hormone-Binding Globulin: Marker of Inflammation? Clin. Chim. Acta 2012, 413, 803–807. [Google Scholar] [CrossRef]
- Liu, N.; Feng, Y.; Luo, X.; Ma, X.; Ma, F. Association between Dietary Inflammatory Index and Sex Hormone Binding Globulin and Sex Hormone in U.S. Adult Females. Front. Public Health 2022, 10, 802945. [Google Scholar] [CrossRef] [PubMed]
- Triggianese, P.; Perricone, C.; Chimenti, M.S.; De Carolis, C.; Perricone, R. Innate Immune System at the Maternal-Fetal Interface: Mechanisms of Disease and Targets of Therapy in Pregnancy Syndromes. Am. J. Reprod. Immunol. 2016, 76, 245–257. [Google Scholar] [CrossRef]
- Girardi, G. Complement Activation, a Threat to Pregnancy. Semin Immunopathol. 2018, 40, 103–111. [Google Scholar] [CrossRef]
- Chao, J.; Guo, Y.; Chao, L. Protective Role of Endogenous Kallistatin in Vascular Injury and Senescence by Inhibiting Oxidative Stress and Inflammation. Oxid. Med. Cell Longev. 2018, 2018, 4138560. [Google Scholar] [CrossRef]
- Lee, K.-N.; Cho, I.; Im, E.M.; Oh, E.; Park, K.H. Plasma IGFBP-1, Fas, Kallistatin, and P-Selectin as Predictive Biomarkers of Histologic Chorioamnionitis and Associated Intra-Amniotic Infection in Women with Preterm Labor. Am. J. Reprod. Immunol. 2023, 89, e13645. [Google Scholar] [CrossRef]
- Cho, I.; Lee, K.-N.; Joo, E.; Kim, Y.M.; Kim, T.E.; Park, K.H. Plasma E-Selectin and Kallistatin as Predictive Markers of Histologic Chorioamnionitis in Women with Preterm Premature Rupture of Membranes. Am. J. Reprod. Immunol. 2022, 88, e13584. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Song, Z.; Liu, A.; Dahmen, U.; Yang, X.; Fang, H. Effects of Lipopolysaccharide-Binding Protein (LBP) Single Nucleotide Polymorphism (SNP) in Infections, Inflammatory Diseases, Metabolic Disorders and Cancers. Front. Immunol. 2021, 12, 681810. [Google Scholar] [CrossRef] [PubMed]
- Blairon, L.; Wittebole, X.; Laterre, P.-F. Lipopolysaccharide-Binding Protein Serum Levels in Patients with Severe Sepsis Due to Gram-Positive and Fungal Infections. J. Infect Dis. 2003, 187, 287–291. [Google Scholar] [CrossRef]
- Cheng, J.; Ji, D.; Yin, Y.; Wang, S.; Song, K.; Pan, Q.; Zhang, Q.; Yang, L. Proteomic Profiling of Serum Small Extracellular Vesicles Reveals Immune Signatures of Children with Pneumonia. Transl. Pediatr. 2022, 11, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, N.M.; Salem, I.; Harris, M.A.; Languino, L.R. IFIT3 (Interferon Induced Protein with Tetratricopeptide Repeats 3) Modulates STAT1 Expression in Small Extracellular Vesicles. Biochem. J. 2021, 478, 3905–3921. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.V.; Jeckel, K.M. Placental Lactogen (CSH). In Encyclopedia of Reproduction, 2nd ed.; Skinner, M.K., Ed.; Academic Press: Oxford, UK, 2018; pp. 502–507. ISBN 978-0-12-815145-7. [Google Scholar]
- Jeckel, K.M.; Boyarko, A.C.; Bouma, G.J.; Winger, Q.A.; Anthony, R.V. Chorionic Somatomammotropin Impacts Early Fetal Growth and Placental Gene Expression. J. Endocrinol. 2018, 237, 301–310. [Google Scholar] [CrossRef]
- Kolluri, A.; Ho, M. The Role of Glypican-3 in Regulating Wnt, YAP, and Hedgehog in Liver Cancer. Front. Oncol. 2019, 9, 708. [Google Scholar] [CrossRef] [PubMed]
- Filmus, J.; Selleck, S.B. Glypicans: Proteoglycans with a Surprise. J. Clin. Investig. 2001, 108, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Song, H.H.; Filmus, J. The Role of Glypicans in Mammalian Development. Biochim. Biophys. Acta 2002, 1573, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadou, F.; Phocas, I.; Mantzavinos, T.; Sarandakou, A.; Rizos, D.; Zourlas, P.A. Discordant Secretion of Pregnancy Specific Beta 1-Glycoprotein and Human Chorionic Gonadotropin by Human Pre-Embryos Cultured in Vitro. Fertil. Steril. 1992, 57, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Dekel, N.; Gnainsky, Y.; Granot, I.; Racicot, K.; Mor, G. The Role of Inflammation for a Successful Implantation. Am. J. Reprod. Immunol. 2014, 72, 141–147. [Google Scholar] [CrossRef]
- Ha, C.T.; Wu, J.A.; Irmak, S.; Lisboa, F.A.; Dizon, A.M.; Warren, J.W.; Ergun, S.; Dveksler, G.S. Human Pregnancy Specific Beta-1-Glycoprotein 1 (PSG1) Has a Potential Role in Placental Vascular Morphogenesis1. Biol. Reprod. 2010, 83, 27–35. [Google Scholar] [CrossRef] [PubMed]
- He, Y.-D.; Xu, B.-N.; Song, D.; Wang, Y.-Q.; Yu, F.; Chen, Q.; Zhao, M.-H. Normal Range of Complement Components during Pregnancy: A Prospective Study. Am. J. Reprod. Immunol. 2020, 83, e13202. [Google Scholar] [CrossRef]
- Son, M.; Diamond, B.; Santiago-Schwarz, F. Fundamental Role of C1q in Autoimmunity and Inflammation. Immunol. Res. 2015, 63, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.C.; Sim, R.B.; Lea, S.M.; Fremeaux-Bacchi, V.; Blom, A.M. Complement Factor I in Health and Disease. Mol. Immunol. 2011, 48, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Roversi, P.; Johnson, S.; Caesar, J.J.E.; McLean, F.; Leath, K.J.; Tsiftsoglou, S.A.; Morgan, B.P.; Harris, C.L.; Sim, R.B.; Lea, S.M. Structural Basis for Complement Factor I Control and Its Disease-Associated Sequence Polymorphisms. Proc. Natl. Acad. Sci. USA 2011, 108, 12839–12844. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; Cheng, G. Role of Interleukin 10 Transcriptional Regulation in Inflammation and Autoimmune Disease. Crit. Rev. Immunol. 2012, 32, 23–63. [Google Scholar] [CrossRef] [PubMed]
- Swaim, C.D.; Scott, A.F.; Canadeo, L.A.; Huibregtse, J.M. Extracellular ISG15 Signals Cytokine Secretion Through the LFA-1 Integrin Receptor. Mol. Cell 2017, 68, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Lozovyy, V.; Richardson, L.; Saade, G.; Menon, R. Progesterone Receptor Membrane Components: Key Regulators of Fetal Membrane Integrity. Biol. Reprod. 2020, 104, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.-H.; Lu, A.-C.; Wang, Y.-C.; Cheng, H.-C.; Chang, C.; Chen, P.-H.; Yu, J.-Y.; Fann, M.-J. Protogenin Defines a Transition Stage during Embryonic Neurogenesis and Prevents Precocious Neuronal Differentiation. J. Neurosci. 2010, 30, 4428–4439. [Google Scholar] [CrossRef] [PubMed]
- Vogel, W.F.; Aszódi, A.; Alves, F.; Pawson, T. Discoidin Domain Receptor 1 Tyrosine Kinase Has an Essential Role in Mammary Gland Development. Mol. Cell Biol. 2001, 21, 2906–2917. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Niswander, L.A. TMEM132A, a Novel Wnt Signaling Pathway Regulator Through Wntless (WLS) Interaction. Front. Cell Dev. Biol. 2020, 8, 599890. [Google Scholar] [CrossRef]
- Maluenda, J.; Manso, C.; Quevarec, L.; Vivanti, A.; Marguet, F.; Gonzales, M.; Guimiot, F.; Petit, F.; Toutain, A.; Whalen, S.; et al. Mutations in GLDN, Encoding Gliomedin, a Critical Component of the Nodes of Ranvier, Are Responsible for Lethal Arthrogryposis. Am. J. Hum. Genet 2016, 99, 928–933. [Google Scholar] [CrossRef]
- Pasterkamp, R.J.; Peschon, J.J.; Spriggs, M.K.; Kolodkin, A.L. Semaphorin 7A Promotes Axon Outgrowth through Integrins and MAPKs. Nature 2003, 424, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Omori, Y.; Katoh, K.; Kondo, M.; Kanagawa, M.; Miyata, K.; Funabiki, K.; Koyasu, T.; Kajimura, N.; Miyoshi, T.; et al. Pikachurin, a Dystroglycan Ligand, Is Essential for Photoreceptor Ribbon Synapse Formation. Nat. Neurosci. 2008, 11, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, D.; Kancharla, S.; Kolli, P.; Sharma, A.K.; Singh, S.; Kumar, S.; Mohanty, A.K.; Jena, M.K. Role of Fibulins in Embryonic Stage Development and Their Involvement in Various Diseases. Biomolecules 2021, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C.; Ahluwalia, N.; Albers, R.; Bosco, N.; Bourdet-Sicard, R.; Haller, D.; Holgate, S.T.; Jönsson, L.S.; Latulippe, M.E.; Marcos, A.; et al. A Consideration of Biomarkers to Be Used for Evaluation of Inflammation in Human Nutritional Studies. Br. J. Nutr. 2013, 109, S1–S34. [Google Scholar] [CrossRef] [PubMed]


| Proteomics Substudy (N = 435) | Not Selected (N = 973) | |
|---|---|---|
| Randomized supplement allocation | ||
| Multiple Micronutrients | 210 (48.3%) | 470 (48.3%) |
| Iron–Folic Acid | 225 (51.7%) | 503 (51.7%) |
| Education | ||
| No schooling | 100 (23.0%) | 258 (26.5%) |
| Grade 1 to Grade 4 | 65 (14.9%) | 116 (11.9%) |
| Grade 5 to Grade 9 | 240 (55.2%) | 509 (52.3%) |
| 10 years and above | 30 (6.9%) | 90 (9.3%) |
| Age (years) | ||
| <20 | 147 (33.8%) | 304 (31.2%) |
| 20–29 | 240 (55.2%) | 546 (56.1%) |
| ≥30 | 48 (11.0%) | 123 (12.6%) |
| Parity | ||
| 0 | 167 (38.4%) | 384 (39.4%) |
| 1 | 139 (31.9%) | 309 (31.8%) |
| ≥2 | 129 (29.6%) | 280 (28.8%) |
| Gestational age at blood draw (wk), median (IQR) | 9.7 (8.0, 12.3) | 10.1 (8.3, 13.4) |
| Maternal anthropometry, mean (SD) | ||
| Height, cm | 149.24 (5.18) | 149.0 (5.3) |
| Weight, kg, | 42.37 (5.76) | 42.9 (6.2) |
| BMI, kg/m2 | 18.99 (2.11) | 19.3 (2.3) |
| Maternal malnutrition | ||
| BMI < 18.5 kg/m2 | 194 (44.6%) | 398 (41.0%) |
| Height < 150 cm, % | 228 (52.4%) | 547 (56.3%) |
| Mid-upper arm circumference <21.5 cm, % | 72 (16.6%) | 178 (18.3%) |
| Living standard index, median (IQR) | −0.22 (−0.70, 0.43) | −0.20 (−0.70, 0.67) |
| Laboratory values | ||
| α-1 acid glycoprotein, g/L | 0.81 (0.30) | 0.73 (0.28) |
| C-reactive protein, mg/L2 | 2.24 (5.25) | NA |
| α-1 acid glycoprotein >1 g/L | 85 (19.5) | 138 (14.2) |
| C-reactive protein >5 mg/L, % 2 | 43 (11.4) | NA |
| Morbidity symptoms (days/wk, yes, %) | ||
| Nausea | 224 (51.5%) | 446 (45.8%) |
| Vomiting | 117 (26.9%) | 237 (24.4%) |
| Severe headache | 31 (7.1%) | 80 (8.2%) |
| High fever | 6 (1.4%) | 20 (2.1%) |
| Low fever | 133 (30.6%) | 310 (31.9%) |
| Diarrhea | 2 (0.5%) | 7 (0.7%) |
| Entrez Gene Symbol | Protein Full Name | Entrez Gene ID | UniProt ID | Pearson r | Abs Δ/2x Protein | Standard Error | p Value | Q Value (FDR) |
|---|---|---|---|---|---|---|---|---|
| SERPINA4 | Kallistatin #‡ | 5267 | P29622 | −0.402 | −0.630 | 0.069 | 2.49 × 10−18 | 1.89 × 10−14 |
| APCS | Serum amyloid P-component # | 325 | P02743 | 0.374 | 0.416 | 0.050 | 7.31 × 10−16 | 1.85 × 10−12 |
| C4BPA | C4b-binding protein alpha chain # | 722 | P04003 | 0.342 | 0.398 | 0.053 | 2.38 × 10−13 | 4.53 × 10−10 |
| SAA1 | Serum amyloid A-1 protein #‡ | 6288 | P0DJI8 | 0.334 | 0.077 | 0.010 | 9.14 × 10−13 | 1.39 × 10−9 |
| SAA2 | Serum amyloid A-2 protein # | 6289 | P0DJI9 | 0.307 | 0.164 | 0.024 | 6.23 × 10−11 | 7.89 × 10−8 |
| ARL5B | ADP-ribosylation factor-like protein 5B # | 221079 | Q96KC2 | 0.302 | 0.433 | 0.066 | 1.27 × 10−10 | 1.38 × 10−7 |
| C9 | Complement component C9 #‡ | 735 | P02748 | 0.289 | 0.376 | 0.060 | 8.37 × 10−10 | 7.95 × 10−7 |
| AFM | Afamin #‡ | 173 | P43652 | −0.288 | −0.354 | 0.057 | 9.88 × 10−10 | 8.34 × 10−7 |
| CCN5 | WNT1-inducible-signaling pathway protein 2 # | 8839 | O76076 | −0.281 | −0.188 | 0.031 | 2.31 × 10−9 | 1.75 × 10−6 |
| RBP4 | Retinol-binding protein 4 ‡ | 5950 | P02753 | −0.276 | −0.292 | 0.049 | 4.94 × 10−9 | 3.41 × 10−6 |
| TFF3 | Trefoil factor 3 #‡ | 7033 | Q07654 | −0.273 | −0.074 | 0.013 | 6.85 × 10−9 | 4.34 × 10−6 |
| SHBG | Sex hormone-binding globulin #‡ | 6462 | P04278 | −0.272 | −0.106 | 0.018 | 8.43 × 10−9 | 4.93 × 10−6 |
| CRP | C-reactive protein # | 1401 | P02741 | 0.270 | 0.051 | 0.009 | 1.04 × 10−8 | 5.65 × 10−6 |
| ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 # | 3698 | P19823 | −0.268 | −0.424 | 0.073 | 1.45 × 10−8 | 6.89 × 10−6 |
| HSPA1A | Heat shock 70 kDa protein 1A # | 3303 | P0DMV8 | −0.263 | −0.275 | 0.049 | 2.54 × 10−8 | 1.13 × 10−5 |
| AHSG | Alpha-2-HS-glycoprotein | 197 | P02765 | −0.259 | −0.359 | 0.064 | 4.43 × 10−8 | 1.87 × 10−5 |
| C1QC | Complement C1q subcomponent subunit C # | 714 | P02747 | 0.250 | 0.098 | 0.018 | 1.21 × 10−7 | 4.16 × 10−5 |
| CFB | Complement factor B # | 629 | P00751 | 0.251 | 0.403 | 0.075 | 1.17 × 10−7 | 4.16 × 10−5 |
| ITIH5 | Inter-alpha-trypsin inhibitor heavy chain H5 | 80760 | Q86UX2 | −0.251 | −0.141 | 0.026 | 1.19 × 10−7 | 4.16 × 10−5 |
| CFI | Complement factor I # | 3426 | P05156 | 0.246 | 0.513 | 0.097 | 1.96 × 10−7 | 6.49 × 10−5 |
| CEP76 | Centrosomal protein of 76 kDa | 79959 | Q8TAP6 | 0.245 | 0.229 | 0.044 | 2.21 × 10−7 | 7.01 × 10−5 |
| BTD | Biotinidase # | 686 | P43251 | −0.244 | −0.372 | 0.071 | 2.44 × 10−7 | 7.42 × 10−5 |
| MFAP2 | Microfibrillar-associated protein 2 | 4237 | P55001 | −0.235 | −0.246 | 0.049 | 7.32 × 10−7 | 2.06 × 10−4 |
| LIPG | Endothelial cell-derived lipase # | 9388 | Q9Y5X9 | 0.234 | 0.217 | 0.043 | 7.73 × 10−7 | 2.10 × 10−4 |
| SERPINA11 | Serpin A11 | 256394 | Q86U17 | −0.224 | −0.171 | 0.036 | 2.39 × 10−6 | 5.86 × 10−4 |
| HYAL1 | Hyaluronidase-1 | 3373 | Q12794 | −0.221 | −0.200 | 0.042 | 3.10 × 10−6 | 7.13 × 10−4 |
| FBLN7 | Fibulin-7 | 129804 | Q53RD9 | −0.219 | −0.186 | 0.040 | 4.13 × 10−6 | 9.23 × 10−4 |
| DDR1 | Epithelial discoidin domain-containing receptor 1 | 780 | Q08345 | −0.218 | −0.176 | 0.038 | 4.64 × 10−6 | 9.52 × 10−4 |
| SLC5A8 | Sodium-coupled monocarboxylate transporter 1 | 160728 | Q8N695 | −0.218 | −0.186 | 0.040 | 4.52 × 10−6 | 9.52 × 10−4 |
| IGSF3 | Immunoglobulin superfamily member 3 | 3321 | O75054 | −0.218 | −0.121 | 0.026 | 4.44 × 10−6 | 9.52 × 10−4 |
| SERPINA7 | Thyroxine-binding globulin # | 6906 | P05543 | −0.216 | −0.340 | 0.074 | 5.22 × 10−6 | 1.04 × 10−3 |
| IFIT3 | Interferon-induced protein with tetratricopeptide repeats 3 | 3437 | O14879 | 0.215 | 0.088 | 0.019 | 5.92 × 10−6 | 1.15 × 10−3 |
| PEF1 | Peflin | 553115 | Q9UBV8 | −0.214 | −0.242 | 0.053 | 6.53 × 10−6 | 1.24 × 10−3 |
| TMEM132A | Transmembrane protein 132A | 54972 | Q24JP5 | −0.214 | −0.115 | 0.025 | 6.69 × 10−6 | 1.24 × 10−3 |
| F11 | Coagulation Factor XI | 2160 | P03951 | 0.213 | 0.215 | 0.048 | 7.60 × 10−6 | 1.35 × 10−3 |
| SERPINA3 | Alpha-1-antichymotrypsin complex | 12 | P01011 | 0.210 | 0.121 | 0.027 | 1.05 × 10−5 | 1.82 × 10−3 |
| MLYCD | Malonyl-CoA decarboxylase, mitochondrial | 23417 | O95822 | 0.208 | 0.353 | 0.080 | 1.17 × 10−5 | 1.89 × 10−3 |
| TLR5 | Toll-like receptor 5 | 7100 | O60602 | 0.208 | 0.223 | 0.050 | 1.17 × 10−5 | 1.89 × 10−3 |
| AGT | Angiotensinogen # | 183 | P01019 | −0.209 | −0.107 | 0.024 | 1.16 × 10−5 | 1.89 × 10−3 |
| MX1 | Interferon-induced GTP-binding protein Mx1 | 4599 | P20591 | 0.208 | 0.062 | 0.014 | 1.19 × 10−5 | 1.89 × 10−3 |
| ISG15 | Ubiquitin-like protein ISG15 | 9636 | P05161 | 0.207 | 0.116 | 0.026 | 1.35 × 10−5 | 2.09 × 10−3 |
| SELPLG | P-selectin glycoprotein ligand 1 # | 6404 | Q14242 | 0.200 | 0.247 | 0.058 | 2.55 × 10−5 | 3.80 × 10−3 |
| PLA2G7 | Platelet-activating factor acetylhydrolase | 7941 | Q13093 | −0.198 | −0.139 | 0.033 | 3.13 × 10−5 | 4.49 × 10−3 |
| C1QL2 | Complement C1q-like protein 2 | 165257 | Q7Z5L3 | −0.198 | −0.191 | 0.045 | 3.11 × 10−5 | 4.49 × 10−3 |
| SERPING1 | Plasma protease C1 inhibitor | 710 | P05155 | 0.197 | 0.182 | 0.043 | 3.37 × 10−5 | 4.74 × 10−3 |
| PCYOX1 | Prenylcysteine oxidase 1 | 51449 | Q9UHG3 | −0.197 | −0.167 | 0.040 | 3.46 × 10−5 | 4.78 × 10−3 |
| APOA1 | Apolipoprotein A-I | 335 | P02647 | −0.196 | −0.143 | 0.034 | 3.71 × 10−5 | 5.04 × 10−3 |
| PGRMC2 | Membrane-associated progesterone receptor component 2 | 10424 | O15173 | −0.196 | −0.077 | 0.019 | 3.78 × 10−5 | 5.04 × 10−3 |
| BGN | Biglycan | 633 | P21810 | −0.195 | −0.100 | 0.024 | 4.08 × 10−5 | 5.34 × 10−3 |
| HS6ST1 | Heparan-sulfate 6-O-sulfotransferase 1 | 9394 | O60243 | −0.195 | −0.246 | 0.060 | 4.35 × 10−5 | 5.60 × 10−3 |
| Entrez Gene Symbol | Protein Full Name | Entrez Gene ID | UniProt ID | Pearson r | Percent Δ/2x Protein | Standard Error | p Value | Q Value (FDR) |
|---|---|---|---|---|---|---|---|---|
| CRP | C-reactive protein # | 1401 | P02741 | 0.894 | 1.190 | 0.029 | 1.16 × 10−147 | 8.48 × 10−144 |
| SAA1 | Serum amyloid A-1 protein #‡ | 6288 | P0DJI8 | 0.654 | 1.067 | 0.060 | 1.57 × 10−52 | 5.72 × 10−49 |
| SAA2 | Serum amyloid A-2 protein # | 6289 | P0DJI9 | 0.556 | 2.086 | 0.152 | 1.65 × 10−35 | 4.02 × 10−32 |
| CLPSL1 | Colipase-like protein 1 | 340204 | A2RUU4 | 0.544 | 2.878 | 0.217 | 9.92 × 10−34 | 1.81 × 10−30 |
| TXNL4A | Thioredoxin-like protein 4A | 10907 | P83876 | 0.524 | 2.140 | 0.170 | 5.09 × 10−31 | 7.42 × 10−28 |
| LBP | Lipopolysaccharide-binding protein # | 3929 | P18428 | 0.515 | 2.052 | 0.167 | 7.85 × 10−30 | 9.54 × 10−27 |
| KIF3A | Kinesin-like protein KIF3A # | 11127 | Q9Y496 | 0.511 | 4.759 | 0.391 | 2.24 × 10−29 | 2.34 × 10−26 |
| C9 | Complement component C9 #‡ | 735 | P02748 | 0.480 | 4.432 | 0.396 | 1.39 × 10−25 | 1.26 × 10−22 |
| APCS | Serum amyloid P-component # | 325 | P02743 | 0.469 | 3.667 | 0.338 | 2.33 × 10−24 | 1.89 × 10−21 |
| SULT2B1 | Sulfotransferase family cytosolic 2B member 1 # | 6820 | O00204 | 0.445 | 4.480 | 0.441 | 8.35 × 10−22 | 6.08 × 10−19 |
| PGLYRP2 | N-acetylmuramoyl-L-alanine amidase | 114770 | Q96PD5 | −0.418 | −3.219 | 0.342 | 3.24 × 10−19 | 2.15 × 10−16 |
| ALB | Serum albumin # | 213 | P02768 | −0.417 | −4.118 | 0.439 | 4.48 × 10−19 | 2.51 × 10−16 |
| CFB | Complement factor B # | 629 | P00751 | 0.417 | 4.766 | 0.508 | 4.35 × 10−19 | 2.51 × 10−16 |
| PLA2G2A | Phospholipase A2, membrane associated # | 5320 | P14555 | 0.409 | 2.242 | 0.245 | 2.47 × 10−18 | 1.29 × 10−15 |
| SAA1 | Serum amyloid A-1 protein # | 6288 | P0DJI8 | 0.404 | 0.597 | 0.066 | 6.30 × 10−18 | 3.06 × 10−15 |
| SERPINA4 | Kallistatin #‡ | 5267 | P29622 | −0.401 | −4.030 | 0.450 | 1.10 × 10−17 | 5.01 × 10−15 |
| LRIG1 | Leucine-rich repeats and immunoglobulin-like domains protein 1 | 26018 | Q96JA1 | 0.395 | 2.410 | 0.274 | 4.14 × 10−17 | 1.78 × 10−14 |
| CLEC3B | Tetranectin | 7123 | P05452 | −0.389 | −3.706 | 0.430 | 1.32 × 10−16 | 5.35 × 10−14 |
| CFI | Complement factor I # | 3426 | P05156 | 0.371 | 5.489 | 0.672 | 3.68 × 10−15 | 1.34 × 10−12 |
| LIPG | Endothelial cell-derived lipase # | 9388 | Q9Y5X9 | 0.368 | 2.414 | 0.298 | 6.68 × 10−15 | 2.21 × 10−12 |
| FGL1 | Fibrinogen-like protein 1 | 2267 | Q08830 | 0.364 | 1.302 | 0.163 | 1.33 × 10−14 | 4.21 × 10−12 |
| LSAMP | Limbic system-associated membrane protein | 4045 | Q13449 | −0.357 | −3.273 | 0.418 | 4.25 × 10−14 | 1.29 × 10−11 |
| HTRA1 | Serine protease HTRA1 | 5654 | Q92743 | 0.345 | 2.054 | 0.273 | 3.46 × 10−13 | 1.01 × 10−10 |
| GDF11|MSTN | Growth/differentiation factor 11/8 | 10220|2660 | O95390|O14793 | −0.343 | −2.122 | 0.284 | 4.68 × 10−13 | 1.31 × 10−10 |
| F9 | Coagulation factor IX | 2158 | P00740 | 0.340 | 3.971 | 0.537 | 7.50 × 10−13 | 2.02 × 10−10 |
| TNXB | Tenascin-X | 7148 | P22105 | −0.334 | −2.088 | 0.288 | 2.05 × 10−12 | 5.34 × 10−10 |
| PLBD1 | Phospholipase B-like 1 | 79887 | Q6P4A8 | 0.332 | 2.013 | 0.280 | 2.77 × 10−12 | 6.95 × 10−10 |
| AOC3 | Vascular adhesion protein-1 | 8639 | Q16853 | −0.328 | −2.235 | 0.315 | 5.76 × 10−12 | 1.40 × 10−9 |
| FGFR1 | Fibroblast growth factor receptor 1 | 2260 | P11362 | −0.323 | −2.647 | 0.379 | 1.17 × 10−11 | 2.74 × 10−9 |
| LUM | Lumican | 4060 | P51884 | −0.322 | −2.523 | 0.363 | 1.42 × 10−11 | 3.24 × 10−9 |
| RET | Proto-oncogene tyrosine-protein kinase receptor Ret | 5979 | P07949 | 0.321 | 0.997 | 0.144 | 1.54 × 10−11 | 3.40 × 10−9 |
| CFHR5 | Complement factor H-related protein 5 ‡ | 81494 | Q9BXR6 | 0.319 | 1.558 | 0.227 | 2.31 × 10−11 | 4.95 × 10−9 |
| NPW | Neuropeptide W | 283869 | Q8N729 | −0.318 | −0.777 | 0.113 | 2.38 × 10−11 | 4.95 × 10−9 |
| NTM | Neurotrimin ‡ | 50863 | Q9P121 | −0.318 | −2.093 | 0.305 | 2.46 × 10−11 | 4.97 × 10−9 |
| NEO1 | Neogenin | 4756 | Q92859 | −0.318 | −2.476 | 0.361 | 2.53 × 10−11 | 4.99 × 10−9 |
| CNTN1 | Contactin-1 | 1272 | Q12860 | −0.312 | −2.277 | 0.340 | 6.60 × 10−11 | 1.23 × 10−8 |
| ALDH7A1 | Alpha-aminoadipic semialdehyde dehydrogenase | 501 | P49419 | 0.310 | 1.588 | 0.238 | 8.09 × 10−11 | 1.47 × 10−8 |
| SPON1 | Spondin-1 | 10418 | Q9HCB6 | −0.309 | −2.860 | 0.430 | 9.20 × 10−11 | 1.64 × 10−8 |
| CHAD | Chondroadherin | 1101 | O15335 | −0.309 | −1.115 | 0.168 | 1.00 × 10−10 | 1.73 × 10−8 |
| CFHR1 | Complement factor H-related protein 1 | 3078 | Q03591 | 0.305 | 3.337 | 0.510 | 1.72 × 10−10 | 2.84 × 10−8 |
| APOA5 | Apolipoprotein A-V | 116519 | Q6Q788 | 0.304 | 1.032 | 0.158 | 1.89 × 10−10 | 3.00 × 10−8 |
| HS6ST3 | Heparan-sulfate 6-O-sulfotransferase 3 | 266722 | Q8IZP7 | −0.304 | −2.697 | 0.413 | 1.90 × 10−10 | 3.00 × 10−8 |
| TP53 | Cellular tumor antigen p53 R175H mutant | 7157 | P04637 | 0.302 | 2.851 | 0.440 | 2.61 × 10−10 | 4.04 × 10−8 |
| C2 | Complement C2 | 717 | P06681 | 0.301 | 3.094 | 0.479 | 2.99 × 10−10 | 4.54 × 10−8 |
| CHL1 | Neural cell adhesion molecule L1-like protein | 10752 | O00533 | −0.300 | −2.903 | 0.451 | 3.27 × 10−10 | 4.87 × 10−8 |
| CFHR5 | Complement factor H-related protein 5 | 81494 | Q9BXR6 | 0.300 | 1.707 | 0.265 | 3.39 × 10−10 | 4.94 × 10−8 |
| TFF3 | Trefoil factor 3 | 7033 | Q07654 | 0.297 | 0.660 | 0.104 | 5.04 × 10−10 | 7.21 × 10−8 |
| CHL1 | Neural cell adhesion molecule L1-like protein | 10752 | O00533 | −0.296 | −1.841 | 0.291 | 6.08 × 10−10 | 8.52 × 10−8 |
| LILRA5 | Leukocyte immunoglobulin-like receptor subfamily A member 5 | 353514 | A6NI73 | 0.296 | 2.505 | 0.396 | 6.30 × 10−10 | 8.66 × 10−8 |
| Symptoms Reported in the Past 7 Days | |||||||
|---|---|---|---|---|---|---|---|
| Nausea | Vomiting | Severe Headache | High Fever | Low Fever | Diarrhea | ||
| Number of participants reporting symptoms (yes, %) | 224 (51.5) | 117 (26.9) | 31 (7.1) | 6 (1.4) | 133 (30.6) | 2 (0.5) | |
| α-1 acid glycoprotein (AGP) (n = 147 proteins) | |||||||
| PC1 (22% variance explained) | 1.04 (0.97, 1.12) | 1.15 (1.04, 1.28) | 1.17 (0.97, 1.41) | 2.28 (1.59, 3.29) | 1.08 (0.99, 1.17) | 0.66 (0.15, 2.10) | |
| PC2 (9% variance explained) | 1.00 (0.93, 1.08) | 1.05 (0.95, 1.17) | 1.22 (1.01, 1.48) | 2.05 (1.37, 3.11) | 1.06 (0.97, 1.15) | 0.60 (0.18, 1.89) | |
| PC3 (6% variance explained) | 1.29 (1.19, 1.39) | 1.13 (1.01, 1.26) | 0.98 (0.82, 1.19) | 1.69 (1.08, 2.77) | 0.98 (0.90, 1.07) | 0.68 (0.31, 2.01) | |
| C-reactive protein (CRP) (n = 879 proteins) | |||||||
| PC1 (24% variance explained) | 0.97 (0.90, 1.04) | 0.94 (0.85, 1.04) | 1.08 (0.89, 1.31) | 1.15 (0.79, 1.70) | 1.01 (0.92, 1.09) | 0.91 (0.28, 2.89) | |
| PC2 (9% variance explained) | 0.97 (0.91, 1.05) | 1.06 (0.95, 1.17) | 1.22 (1.02, 1.45) | 0.96 (0.64, 1.39) | 1.07 (0.99, 1.17) | 0.59 (0.13, 1.91) | |
| PC3 (5% variance explained) | 1.05 (0.97, 1.12) | 1.19 (1.07, 1.33) | 1.05 (0.87, 1.28) | 2.52 (1.54, 4.33) | 1.01 (0.93, 1.10) | 0.76 (0.27, 2.47) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Bedsaul-Fryer, J.R.; Schulze, K.J.; Sincerbeaux, G.; Baker, S.; Rebholz, C.M.; Wu, L.S.; Gogain, J.; Cuddeback, L.; Yager, J.D.; et al. An Early Gestation Plasma Inflammasome in Rural Bangladeshi Women. Biomolecules 2024, 14, 736. https://doi.org/10.3390/biom14070736
Kim H, Bedsaul-Fryer JR, Schulze KJ, Sincerbeaux G, Baker S, Rebholz CM, Wu LS, Gogain J, Cuddeback L, Yager JD, et al. An Early Gestation Plasma Inflammasome in Rural Bangladeshi Women. Biomolecules. 2024; 14(7):736. https://doi.org/10.3390/biom14070736
Chicago/Turabian StyleKim, Hyunju, Jacquelyn R. Bedsaul-Fryer, Kerry J. Schulze, Gwen Sincerbeaux, Sarah Baker, Casey M. Rebholz, Lee SF Wu, Joseph Gogain, Lena Cuddeback, James D. Yager, and et al. 2024. "An Early Gestation Plasma Inflammasome in Rural Bangladeshi Women" Biomolecules 14, no. 7: 736. https://doi.org/10.3390/biom14070736







