Abstract
Signaling pathways are responsible for transmitting information between cells and regulating cell growth, differentiation, and death. Proteins in cells form complexes by interacting with each other through specific structural domains, playing a crucial role in various biological functions and cell signaling pathways. Protein–protein interactions (PPIs) within cell signaling pathways are essential for signal transmission and regulation. The spatiotemporal features of PPIs in signaling pathways are crucial for comprehending the regulatory mechanisms of signal transduction. Bimolecular fluorescence complementation (BiFC) is one kind of imaging tool for the direct visualization of PPIs in living cells and has been widely utilized to uncover novel PPIs in various organisms. BiFC demonstrates significant potential for application in various areas of biological research, drug development, disease diagnosis and treatment, and other related fields. This review systematically summarizes and analyzes the technical advancement of BiFC and its utilization in elucidating PPIs within established cell signaling pathways, including TOR, PI3K/Akt, Wnt/β-catenin, NF-κB, and MAPK. Additionally, it explores the application of this technology in revealing PPIs within the plant hormone signaling pathways of ethylene, auxin, Gibberellin, and abscisic acid. Using BiFC in conjunction with CRISPR-Cas9, live-cell imaging, and ultra-high-resolution microscopy will enhance our comprehension of PPIs in cell signaling pathways.
1. Introduction
Cellular signaling pathways are intricate biochemical reactions and play a crucial role in facilitating information transmission in living organisms, enabling cells to promptly and accurately respond to external stimuli [1,2,3,4]. The transmission of signals in biological cells is primarily facilitated through molecular interactions, particularly protein–protein interactions (PPI) [5,6,7,8]. The real-time observation of PPI dynamics, alterations, and spatial-temporal distribution is crucial for elucidating the regulatory interplay among various genes. Conventional biochemical techniques utilized for the examination of PPI, such as co-immunoprecipitation (Co-IP) and immunofluorescence (IF), are constrained in their ability to offer insights into the transient and dynamic nature of interactions as well as to provide accurate and detailed visualization. In light of the advancements in molecular biology and biotechnology, researchers have been actively investigating novel approaches to elucidate these intricate signaling pathways. Notably, bimolecular fluorescence complementarity (BiFC) has emerged as a valuable tool to explore protein interactions, based on the division and recombination of fluorescent proteins, which are usually divided into an N-terminal fragment and a C-terminal fragment [9,10]. When two target proteins interact with each other, the N-terminal and C-terminal fragments of the fluorescent protein will approach each other and recombine to restore the original structure and properties, thus generating fluorescence signals to reflect the interaction of the target proteins, offering substantial assistance in the examination of cellular signaling pathways [10,11,12] (Figure 1A). BiFC has significantly contributed to the investigation of cellular signaling pathways by enabling the examination of interactions between signaling proteins and their downstream effector proteins as well as the analysis of signal transmission within cells [13,14,15]. This visualization technique aids in comprehending the composition and structure of signaling pathways as well as in analyzing the dynamic alterations of signaling pathways in various physiological and pathological states [16,17,18,19,20]. Its utility extends to the study of fundamental biological processes including cell signal transduction, gene expression regulation, and protein complex formation [12,21,22]. By conducting comprehensive investigations into these processes, valuable insights into the underlying principles and mechanisms governing life activities can be obtained. Collectively, BiFC allows for the in situ detection of PPIs, real-time monitoring of cellular activity, and examination of subcellular localization, thus offering a distinct viewpoint for uncovering critical regulatory relationships and signaling pathways within gene regulatory networks.
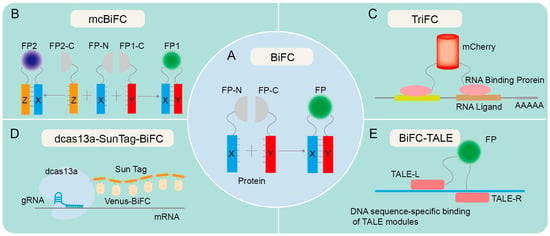
Figure 1.
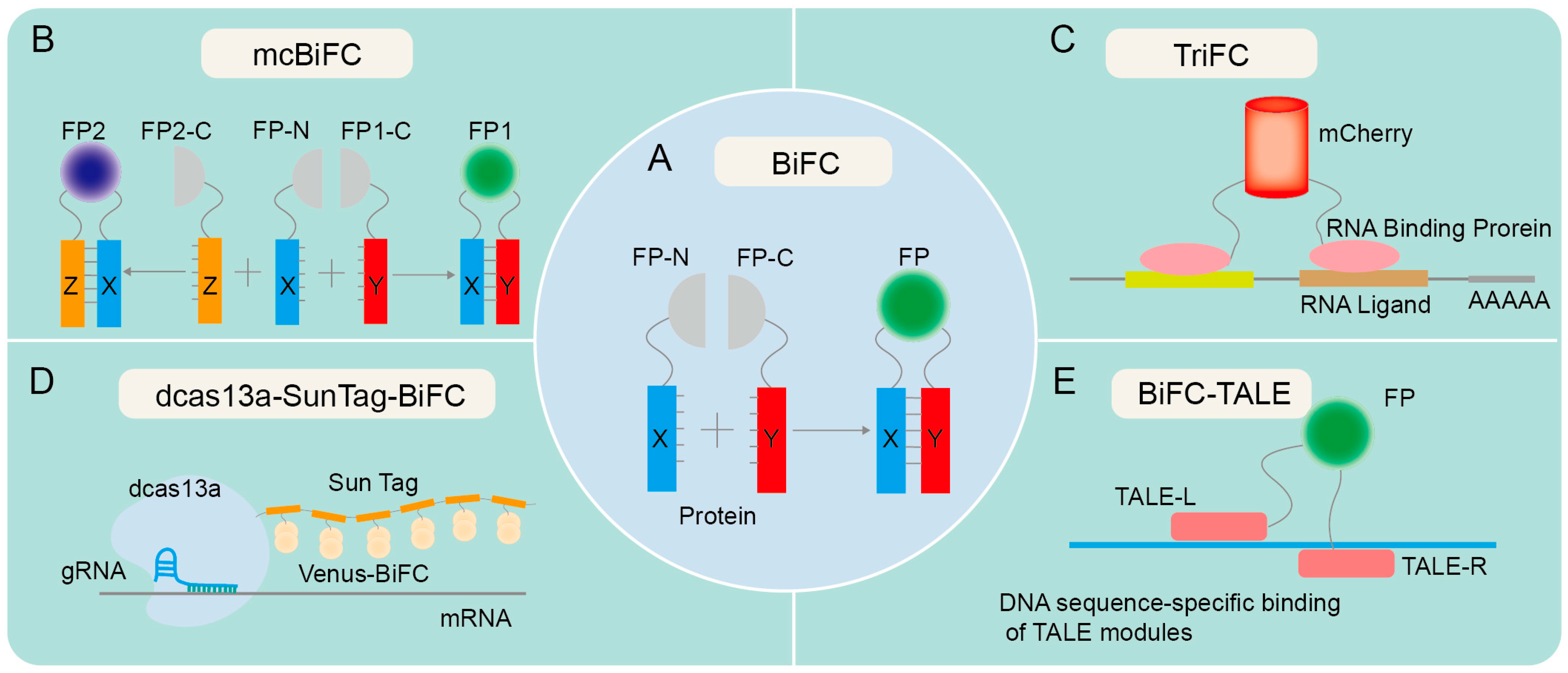
Technical principles of various forms of BiFC. (A) The principle of traditional BiFC. Proteins X and Y contain an N-terminal fragment and a C-terminal fragment, respectively, and are hypothesized to potentially interact with each other. The fusion of X protein and Y protein facilitates the identification of the interaction between X and Y. If X and Y proteins interact, recombination of the fluorescent protein fragments is expected to result in fluorescence emission. (B) The principle of mcBiFC. X protein interacts with Y and Z proteins, respectively, emitting different fluorescence due to the unique fluorescent protein fragments of Y and Z proteins. (C) The principle of TriFC. The N-terminal fragment of mCherry is linked to the target mRNA via an RNA-binding protein, and its C-terminus is linked to a potential RNA-binding protein. Upon interaction between the candidate RNA-binding protein and the target mRNA, the N- and C-terminal fragments of mCherry undergo recombination, resulting in the production of a red fluorescent protein (RFP) signal. (D) The principle of dcas13a-SunTag-BiFC. The fusion of dCas13a with SunTag tags facilitates the recruitment of fluorescent protein fragments, enabling RNA imaging via the targeted binding of guide RNA (gRNA). (E) The principle of BiFC-TALE. Transcription activator-like effector (TALE) can bind to specific DNA sequences and aggregate fluorescent protein fragments for imaging genomic loci.
Since the emergence of BiFC in the 1990s, there have been several notable advancements in this technology. One such advancement is the development of split GFP variants and other fluorescent proteins with improved brightness and photostability, allowing for more sensitive and quantitative detection of PPIs [10,15,23,24,25]. The simultaneous expression of two or more fluorescent proteins in a multicolor bimolecular fluorescence complementation (mcBiFC) system facilitates the concurrent monitoring of multiple protein interactions and allows for the comparison of their interaction efficiencies [26,27,28] (Figure 1B). Another advancement is the use of BiFC in high-throughput screens, enabling the rapid identification of PPIs on a genome-wide scale [29,30,31,32]. BiFC has also been developed to visualize the interaction between RNA and proteins, as evidenced by trimeric fluorescence complementation (TriFC) systems [33] (Figure 1C). TriFC is optimized by utilizing mNeptune’s good optical properties to image RNA–protein interactions in living animals [34]. The integration of dCas13a with SunTag has been utilized to enhance the recruitment of Venus fragments, thereby enabling the real-time visualization of endogenous specific mRNA in cells through the targeted binding of gRNA with cas13a [35] (Figure 1D). The fusion of BiFC with transcription activator-like effectors (TALEs) enables precise labeling and visualization of genomic loci through the specific interaction of TALEs with target DNA sequences [36] (Figure 1E).
BiFC exhibits significant promise in the fields of disease diagnosis and drug screening. By constructing large-scale PPI networks, researchers can screen for key signaling pathways associated with specific diseases and further investigate potential drug targets in these pathways [37,38,39,40,41,42]. BiFC is a valuable tool for screening potential drug molecules and investigating the interactions between drugs and target proteins, thereby expediting the drug development process and enhancing the effectiveness and safety of medications [37,41,43,44,45,46]. BiFC has the capability to identify PPIs associated with various diseases, thereby offering substantial evidence for the early diagnosis and prognosis assessment of such conditions [47,48,49]. Specifically, the identification of distinct PPIs within cancer cells holds promise for the timely detection and treatment of cancer [49]. BiFC also has the potential to facilitate the creation of novel therapeutic approaches centered on protein interactions [50,51,52,53,54]. Through the manipulation of molecules that disrupt pathogenic protein interactions, the progression of diseases can be impeded; conversely, the promotion of beneficial protein interactions can enhance cellular function recovery and regeneration. In conclusion, it is anticipated that BiFC will assume a greater significance in future investigations pertaining to cell signaling pathways, disease diagnosis, and drug screening, as a result of ongoing advancements and innovations in technology. This review aims to explore the recent progress in BiFC, including technical innovations and applications in investigating PPIs in cell signaling pathways.
2. Overview of BiFC Development
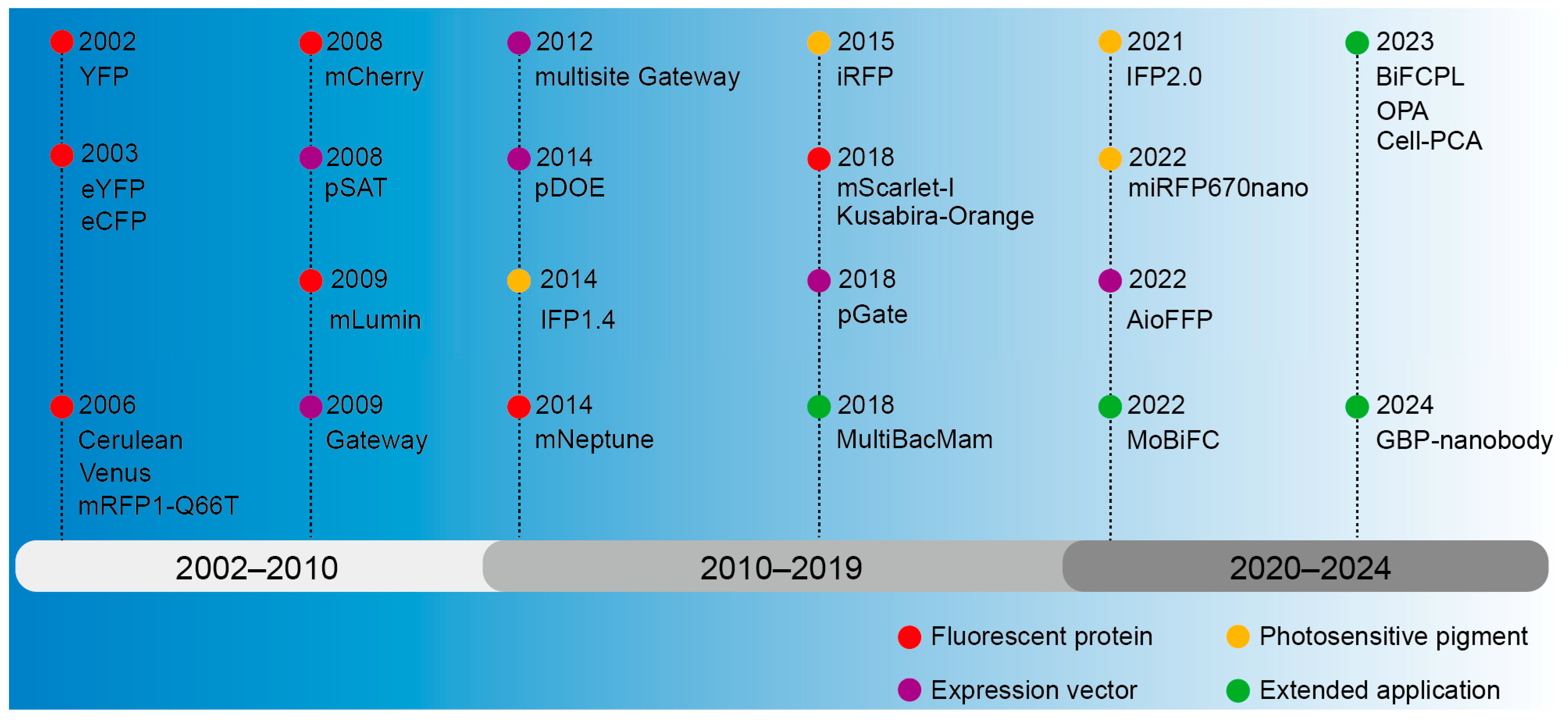
The inception of BiFC technology can be dated back to the early 21st century, when researchers endeavored to develop a dynamic, real-time way to monitor protein interactions [55,56]. The early researchers primarily concentrated on enhancing the selection and cleavage techniques of fluorescent proteins to guarantee the stability and specificity of fluorescent signals [55,57]. Concurrently, they were consistently investigating the utilization of BiFC in various biological systems and cells to broaden its range of applications [56,57]. A ubiquitin-based split-protein sensor (USPS) was introduced in 1994, enabling the real-time monitoring of PPIs within a living cell at their endogenous locations [58]. The USPS assay is capable of detecting a transient in vivo interaction between polypeptides, as exemplified by a close proximity between Sec62p and a nascent polypeptide chain [59]. The use of fluorescent proteins in biological studies has greatly increased since the discovery of Green Fluorescent Protein (GFP) in the 1960s [60]. These important early discoveries laid a solid foundation for the emergence and development of BiFC. The BiFC technology has undergone continuous development over a span of more than two decades since its inception, with a primary emphasis on the enhancement and advancement of fluorescent proteins, photopigments, and expression vectors and an expanding range of applications (Figure 2).
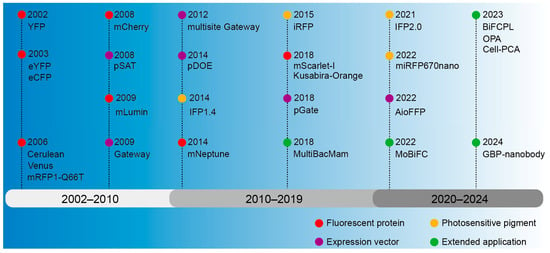
Figure 2.
The development history overview of BiFC.
2.1. Fluorescent Protein Used for Investigating Protein–Protein Interactions
The BiFC technology was first introduced in 2002, employing yellow fluorescent protein (YFP) as a fluorescent marker for investigating PPIs [61]. The GFP gene was first cloned in 1992 from cnidarian, Aequorea victoria [62], and GFP was then confirmed in 2000 to be characteristic of fluorescence complementation [63]. Fluorescent proteins emerged as prominent markers in these early assays owing to their high sensitivity, reliability, and versatility [55,56,57]. In 2003, the multicolor bimolecular fluorescence complementation (mcBiFC) system was developed by using eYFP and eCFP [26]. To enhance fluorescence signals in a physiological culture setting, mutations in Cyan Fluorescent Protein (CFP) and YFP were employed to create Cerulean and Venus variants, and then fluorescence resonance energy transfer with fluorescence lifetime imaging microscopy (FRET-FLIM) was integrated for terrain visualization [64,65]. The development of red fluorescent protein has expanded the scope of BiFC application. The BiFC system based on DsRed variant monomeric RFP (mRFP1-Q66T) [66] and mutant monomeric RFP (mCherry) [67] has a bright red fluorescence excitation and emission wavelength. To facilitate the direct observation of protein interactions in a physiological environment of mammals, a monomeric lumin (mLumin) BiFC system was developed utilizing the mKate variant, enabling the detection of protein interactions at 37 °C and providing a more faithful representation of protein characteristics [68]. To achieve in vivo imaging with an excitation peak above 600 nm, mNeptune was applied to BiFC [34]. The mScarlet-I-based BiFC assay was developed using a traditional β-Fos/β-Jun constitutive heterodimerization model and a rapamycin-inducible FRB/FKBP interaction system, enabling the visualization of diverse PPIs in distinct subcellular compartments with exceptional specificity and sensitivity, particularly at the physiological temperature 37 °C in live mammalian cells [69]. An orange-colored BiFC system was established using the Kusabira-Orange (KO) protein isolated from the stony coral Fungia concinna; as a result, the specificity and sensitivity were enhanced, thus expanding the potential applications of multicolor BiFC analysis [70]. However, the auto-fluorescent protein (AFP) fragment of the BiFC complex tends to accumulate and precipitate in vitro, making it difficult to characterize physical properties, so chimeric AFP was created by directly fusing different DNA encoding AFP fragments [71]. In summary, the localization of proteins within cells and their interactions can be ascertained through fluorescence signals, obviating the necessity for chemical alterations to the proteins.
2.2. Photosensitive Pigments for In Vivo Imaging
Photosensitive pigments are a type of photoreceptor in bacteria or plants that can absorb red and near-infrared light and mediate the transmission of light signals [72]. The iRFP–BiFC system, which utilizes the bacterial photosensitive pigment intrinsic red fluorescent protein (iRFP), emitting long waves, has potential applications for in vivo imaging [73]. A protein-fragment complementation assay (PCA) was introduced based on an engineered Deinococcus radiodurans infrared fluorescent protein IFP1.4, enabling analysis of hormone-induced signaling complexes in living yeast and mammalian cells at the nanometer resolution [74]. Based on cyanobacterial pigments, mini-red fluorescent protein 670 nanoparticles (miRFP670 nano) [75] and IFP2.0 [76] with higher signal intensity and photostability were developed. By activating photosensitive pigments under specific lighting conditions, fluorescence signals can be generated at specific time and spatial points, allowing for the observation of precise regulatory mechanisms for a certain physiological process.
2.3. Vector System Expressing the Fluorescent Fusion Protein
A stable and efficient expression vector of the fusion protein is critical for the BiFC experiments. Expression vectors based on the pSAT series of vectors allow for determining the interaction of a “bait” protein and multiple “prey” proteins and are thus advantageous for the implementation of multicolor BiFC in living plant cells [77]. The Gateway vector system can be applied to screen interacting proteins on a large scale, as different cDNAs can be cloned into vectors without the use of restriction enzymes and ligases [78]. However, the use of multiple vectors will lead to differences in the expression levels of fusion proteins, making it difficult to interpret the interaction results quantitatively, whereas, using the improved Gateway-compatible cloning system, multiple target fragments can be transferred into the same vector to ensure the same amount of expression [79]. Subsequently, a dual-ORF expression BiFC system (pDOE) was constructed, which ensures that the fusion protein can be introduced into transformed cells in a 1:1 concentration ratio, but the efficiency is low when multiple DNA fragments are assembled in a predetermined order [80]. Therefore, the advantages of Golden Gate and Gateway cloning methods were combined to design a set of double-compatible pGate vectors, which could assemble multiple destination sequences in a predetermined order [81]. Furthermore, the creation of an All-in-One fluorescent fusion protein (AioFFP) vector toolbox using Gibson assembly enables the simultaneous incorporation of multiple fluorescent fusion protein expression units into a single plasmid, facilitating the co-expression of multiple genes in BiFC detection [82]. In addition, it is possible to improve the vector by selecting efficient promoters, inserting linker sequences, and optimizing the fusion protein configuration to ensure the robust expression of the fusion protein within the cell.
2.4. The Application of BiFC
The application of BiFC in the field of life science is constantly expanding. To apply BiFC to all mammalian cells and cells that are difficult to transfect with plasmids, the gene delivery tool Multiple Bacullovirus and Mammalian (MultiBacMam) combined with BiFC allows for efficient PPI screening in cells [44]. Through the optimization of the protoplast-based transient expression system (PTES), enhanced efficiency in obtaining active protoplasts can be achieved, and when coupled with the BiFC experiment, this optimized PTES can be effectively utilized for investigating protein interactions in monocotyledonous plants [83]. Modular BiFC (MoBiFC) was developed to observe specific protein interactions in chlorophylls, and its application was extended to other organelles [84]. Organelles perform biological functions through interactions of key proteins in contact with membranes [85]. In 2023, the proximity labeling strategy (BiFCPL) was developed to study the membrane contact proteome between mitochondria and endoplasmic reticulum (MERCs) [86]. In order to further analyze the structure and protein affinity of the complex, One Pattern Analysis (OPA) was recently developed by combining FRET-FLIM and BiFC, which can be used for protein affinity analysis and provide key information such as distance relationship and orientation of PPI [87].
BiFC technology can also be used for large-scale screening of interacting proteins. Lee et al. developed an arrayed screening strategy based on protein complementation, systematically studied protein–protein interactions in living cells, and conducted large-scale screening of telomere regulators, revealing the basic molecular niche of human telomere regulation and providing valuable tools for studying mammalian signaling pathways in cells [88]. Bischof et al. established a multicolor BiFC library, which covers most transcription factors of Drosophila and can be used for large-scale identification and analysis of PPIs in Drosophila [89]. A novel experimental strategy for cell-based protein complementation assay (Cell-PCA) has been proposed, which is based on BiFC, combined with high-throughput sequencing methods, and can be used to screen proteins interacting with different decoy proteins at the whole genome level in the same live-cell context [90]. More recently, a coupled BiFC/GFP binding peptide (GBP) nanobody-based technique was utilized to unravel the function of defined dimers of transcription factors in living cells [91]. In summary, these improvements have diversified and refined the application of BiFC techniques in biological research.
3. Protein–Protein Interactions Revealed by BiFC in Cell Signaling Pathways
3.1. TOR Signaling Pathway
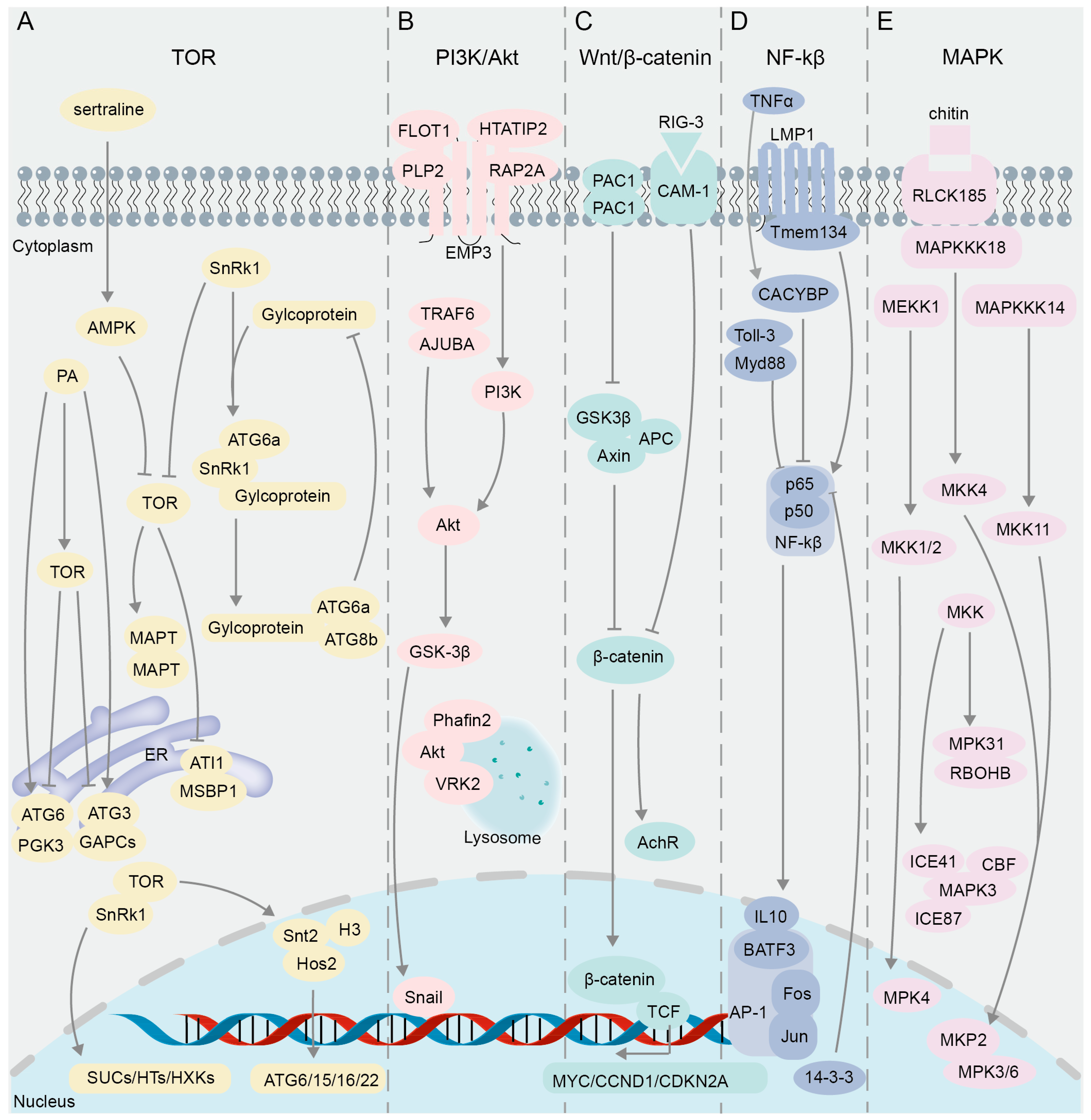
The target of rapamycin (TOR), a serine/threonine kinase within the phosphatidylinositol 3-kinase-related kinase family, governs a signaling network critical for cell cycle progression, protein synthesis, autophagy, and other processes necessary for cell viability and growth (Figure 3A). The TOR signaling pathway is a highly conserved molecular mechanism that regulates cell growth and metabolism in response to environmental and nutritional stimuli. Especially, this pathway regulates intracellular autophagy [92,93,94], which requires the participation of a group of autophagy-related (ATG) proteins [95,96] (Figure 3A). A reduction in TOR activity was proven to trigger the autophagy process, while phosphatidic acid (PA), a negative regulator of autophagy in animals, was found to activate the mammalian target of rapamycin complex 1 (mTORC1) [97]. In the investigation of the suppressive impact of PA on autophagy in plants, multiple assay techniques including BiFC, yeast two-hybrid (Y2H), glutathione S-transferase (GST) pull-down, Co-IP, and FRET-FLIM were used to elucidate the interaction of GAPCs (glyceraldehyde-3-phosphate dehydrogenase), PGK3 (phosphoglycerate kinase 3), and autophagy-related proteins (ATG3 and ATG6) within the endoplasmic reticulum (ER) (Figure 3A), and these complexes were notably strengthened in the presence of PA, suggesting that the interaction complex may play a role in the early regulation of autophagy in the ER [98]. When cells experience disturbances from internal and external stimuli, misfolded proteins accumulate in the ER, triggering the onset of ER autophagy [99]. Through Y2H and BiFC assays, ATG8-interacting proteins 1 and 2 (ATI1 and ATI2) were found to interact directly with AGO1 on the ER in agroinfiltrated tobacco leaves, supporting the involvement of ATI1 and ATI2 in the Argonaute (Ago)-mediated ER autophagy pathway [100]. Y2H and in vivo BiFC assays verified that ER-localized MSBP1 (Membrane Steroid Binding Protein 1) interacts with the selective autophagy cargo receptors ATI1 and ATI2 [101].
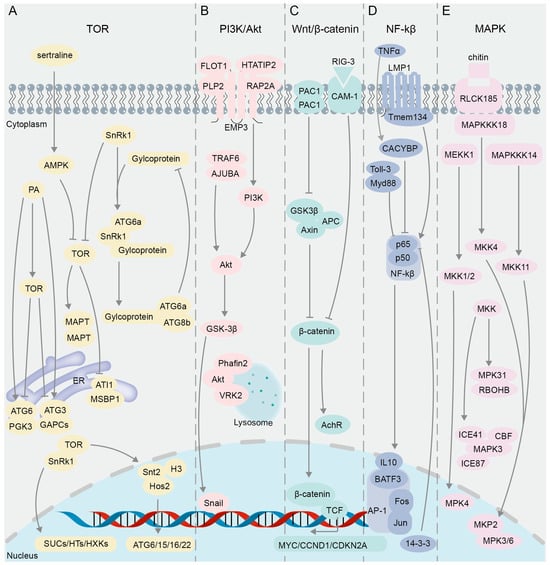
Figure 3.
The interacting proteins in the classical cell signaling pathway validated using BiFC. (A) TOR signaling pathway. (B) PI3K/Akt signaling pathway. (C) Wnt/β-catenin signaling pathway. (D) NF-κB signaling pathway. (E) MAPK signaling pathway.
Post-translational covalent modification of histones is also involved in the regulation of autophagy [102], and mTOR can affect the transcription of histone acetyltransferase and regulate the expression of autophagy genes [103]. By using BiFC and GST pull-down techniques, it was found in Magnaporthe oryzae that the domain PHD (Plant Homeodomain) of Snt2 protein binds to acetylated histone H3, and the domain ELM2 binds to deacetylase Hos2 to regulate the expression of autophagy genes ATG6, 15, 16, and 22, while the expression of Snt2 is positively regulated by TOR [104] (Figure 3A). In response to cellular stress or nutrient deprivation, the sucrose non-fermenting protein-1-related protein kinase (SnRK1) inhibits TOR activity (Figure 3A), leading to the dephosphorylation of ATG13 and its subsequent binding to ATG1 to form precursor complexes, thereby initiating the process of autophagy [96,105,106]. Y2H and BiFC results showed that glycoprotein encoded by Rice stripe mosaic cytorhabdovirus (RSMV) interacts with SnRK1B in Oryza sativa to promote the kinase activity of SnRK1B on ATG6b (Figure 3A), thereby positively regulating autophagy [107]. Studies have shown that SnRK and TOR proteins play critical roles in plant sugar metabolism [108,109]. Results of Y2H and BiFC assays demonstrated that TOR in Vitis vinifera interacts with SnRK1.1 to regulate SUCs (sucrose transporters), HTs (hexose transporters), HXKs (hexokinases), and other glucose metabolism-related genes [110] (Figure 3A).
TOR can also phosphorylate the microtubule-associated protein tau (MAPT), leading to MAPT aggregation and inhibition of autophagy [111]. However, an anti-depressant drug, sertraline (Sert), as an autophagy inducer, can activate AMP-activated protein kinase (AMPK), thereby inhibiting the oligomerization of MAPT in MAPT-BiFC cells [43]. This negative regulation of the TOR pathway promotes the autophagic degradation of MAPT, thus suppressing tauopathy [43]. The subunits TOR1, TOR2, and Kog1 form the TOR Complex 1 (TORC1), which regulates cell growth by modulating autophagy and ribosome biogenesis processes [112,113]. A genome-wide examination of the TORC1 interactome was performed in yeast through BiFC assay, resulting in the identification of predominant BiFC signals localized at the vacuolar membrane, and a subset of these signals were found within cytoplasmic messenger ribonucleoprotein (mRNP) granules, where TORC1 modulates the activity of the translation repressor protein Scd6 via phosphorylation, thereby influencing the regulation of post-transcriptional gene expression [114].
3.2. PI3K/Akt Signaling Pathway
The signaling pathway PI3K/Akt (phosphatidylinositol 3′-kinase and RAC-alpha serine/threonine-protein kinase or protein kinase B) plays a crucial role in intracellular signal transduction (Figure 3B), influencing processes such as cell growth, proliferation, survival, and metabolism [115,116]. In research pertaining to this pathway, the BiFC technique is primarily utilized for the visualization and analysis of PPIs among key components. Snail and GSK-3β, integral constituents of the PI3K/Akt pathway, play a regulatory role in the epithelial–mesenchymal transition (EMT) process [117] (Figure 3B). Results of immunoprecipitation and BiFC assays showed that overexpression of ajuba LIM protein (AJUBA) can recruit tumor necrosis factor associated factor 6 (TRAF6), enhance Akt phosphorylation, activate the Akt/GSK-3β/Snail signaling pathway, and promote the EMT process, thereby enhancing the invasion and metastasis ability of HCC (Hepatocellular Carcinoma cells) [118] (Figure 3B). Epithelial membrane protein 3 (EMP3) can affect the PI3K-Akt pathway in cancer cells [119,120]. Y2H screening identified 10 previously unreported interaction partners of EMP3, eight of which were further verified through BiFC and Proximity Ligation Assay (PLA); among these candidate interactors, Flotillin-1 (FLOT1), HIV Tat-interacting protein 2 (HTATIP2), Ras-related protein 2A (RAP2A), and Proteolipid protein 2 (PLP2) exhibited strong signals in BiFC and PLA assays [121] (Figure 3B). FLOT1 and PLP2 have been identified as positive regulators of the PI3K-AKT signaling pathway, as evidenced by previous studies [122,123]. Conversely, HTATIP2 and CMTM5 (CKLF-like MARVEL transmembrane domain-containing member 5) have been shown to exert negative regulatory effects on this pathway [124,125]. Additionally, RAP2A has been found to exhibit both positive and negative regulatory capabilities in Akt signaling [126,127] (Figure 3B).
Lysosomes are known to be major organelles involved in autophagy [128,129]. Y2H and BiFC assays have substantiated the occurrence of an interaction between Akt and Phafin2 in both the cytoplasm and nucleus; nevertheless, upon treatment of cells with rapamycin or HBSS (Hank’s Balanced Salt Solution), the Akt-Phafin2 complex is augmented in the lysosome, thereby triggering autophagy [130] (Figure 3B). Subsequently, the proteins that interact with the Akt complex in the lysosomes after autophagy induction was further investigated by means of time-of-flight mass spectrometry (TOF/MS), BiFC, and immunofluorescent assays, and it was revealed that VRK2 (Vaccinia-related kinase 2) maintains Akt kinase activity by interacting with Akt in the lysosomes, thereby regulating autophagy and cell proliferation [131] (Figure 3B).
3.3. Wnt/β-Catenin Signaling Pathway
The primary regulator of the Wnt/β-catenin pathway is the Axin/APC/GSK3β destruction complex (DC) (Figure 3C), which allows for the degradation of cytoplasmic β-catenin in the absence of external stimulation. The stability of β-catenin protein is critical in the regulation of the Wnt signaling pathway [132] (Figure 3C). When the Wnt signaling pathway is activated, the Axin/APC/GSK3β destruction complex (DC) becomes inactive, leading to increased stability of its target protein β-catenin, which accumulates and then translocates to the nucleus [133,134,135,136] (Figure 3C). The BiFC assay demonstrated that the conformational alteration of DC is triggered by the presence of Wnt ligands, leading to the inhibition of the Axin–GSK3β interaction in Drosophila, thereby impeding the degradation of β-catenin [137] (Figure 3C). Subsequently, β-catenin and T-cell transcription factor (TCF) interact in the nucleus and synergistically regulate the expression of target genes MYC, CCND1, and CDKN2A to affect cell survival [138] (Figure 3C), and BiFC signals are strong in the S and G2 phases of cells [20]. The dimerization of PAC1 (pituitary adenylate cyclase-activating polypeptide) was confirmed using BiFC and the bioluminescence resonance energy transfer (BRET) assay [139], and the PAC1 dimerization is migrated from the plasma membrane to the nucleus under serum withdrawal conditions [140]. Interestingly, similar to the frizzled receptor dimer [141], the dependent basic activity of PAC1 dimerization can also activate the Wnt/β-catenin pathway, increasing the levels of β-catenin, cyclin D1, and c-myc in the pathway, and as a result, the cells show higher anti-apoptotic activity [140]. In addition, CAM-1, a ROR receptor tyrosine kinase (RTK), is an unconventional receptor of Wnt associated with neural signaling processes dependent on the Wnt pathway [142,143,144]. Through genetic and BiFC assays, it was demonstrated that presynaptic RIG-3, an immunoglobulin superfamily protein, interacts directly with the immunoglobulin domain of postsynaptic CAM-1, a nonconventional Wnt receptor, at the Caenorhabditis elegans neuromuscular junction (NMJ) [145] (Figure 3C). This interaction subsequently suppresses Wnt/LIN-44 signaling, thereby preserving the appropriate levels of acetylcholine receptor, AChR/ ACR-16, at the neuromuscular synapse [145] (Figure 3C).
3.4. NF-κB Signaling Pathway
The NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) family in mammals consists of five closely related transcription factors: p50, p52, p65 (RelA), c-Rel, and RelB [146] (Figure 3D). NF-κB dimers formed by the two subunits p50 and p65 (RelA) are involved in regulating the expression of genes related to cell survival, immunity, and anti-apoptosis [147,148,149]. The activator protein 1 (AP-1) superfamily, as dimeric transcription factors associated with tumor development, is composed of different family protein members such as Jun and Fos [150] (Figure 3D). It was revealed by BiFC that Rel family proteins p50 and p65 in NF-κB interact with Fos and Jun and inhibit their transcriptional activities [61] (Figure 3D). Subsequently, a BiFC-based FRET (BiFC-FRET) assay was used to confirm that the trimer complex formed by the interaction between the p65 and the Fos-Jun heterodimer of AP-1 participates in the regulation of the target gene of AP-1 [65] (Figure 3D). BATF3 is a member of the ATF-like family and belongs to the AP-1 transcription factor family [151,152]. It was proven by BiFC assay that the interaction between CiBATF3 and interleukin 10 (IL-10) in Ctenopharyngodon idella negatively regulates the activity of NF-κB [153] (Figure 3D). GST pull-down and BiFC assays verified that p65 inhibits the activity of NF-κB pathway through interaction with the 14-3-3 proteins in the neuronal nuclei (Figure 3D), thereby protecting neurons from ischemia (I/R) injury and regulating nerve cell survival [154]. By combining BiFC and a transposon gene trap system, researchers have screened the protein Calcyclin Binding Protein (CACYBP) that interacts with p65 and found that their interaction enhances the activity of NF-κB under TNFα stimulation [155] (Figure 3D). Subcellular co-localization and BiFC assay results showed that other protein interaction complexes can also affect the activity of NF-κB. For example, the interaction between LMP1 (latent membrane protein 1) and Tmem134 (transmembrane protein 134) [156] or the interaction between the TIR (Toll-interleukin 1-resistance) domain and Toll-like receptor 3 (Toll-3) [157] can significantly activate the NF-κB pathway and enhance the immune function of the body (Figure 3D).
3.5. MAPK Signaling Pathway
The Mitogen-Activated Protein Kinase (MAPK) signaling pathway, which mainly consists of Mitogen-Activated Protein Kinase Kinase Kinases (MAPKKKs or MEKKs), Mitogen-Activated Protein Kinase Kinases (MAPKKs or MKKs or MEKs), and Mitogen-Activated Protein Kinases (MAPKs or MPKs), regulates a variety of cellular activities including proliferation, differentiation, survival, and death [158] (Figure 3E). This pathway is crucial for regulating immune response mechanisms [159]. Previous studies have shown that MEKK1 and MPK4 are negative regulators of innate immune response in plants, and their deletion can induce constitutive expression of pathogenesis-related genes [160,161], while the MEKK1 mutant can cause programmed cell death [161]. The BiFC assay demonstrated that MEKK1 interacts with MKK1 and MKK2 on the plasma membrane and that the interaction signals of MPK4 with MKK1 and MKK2 appear in the plasma membrane and nucleus [162] (Figure 3E). These results suggest that MEKK1, MKK1/MKK2, and MPK4 can form a kinase cascade to negatively regulate the innate immune response of plants and prevent immune hyperplasia [162]. In mammalian cells, a subfamily of 10 dual-specificity (Thr/Tyr) MAPK phosphatases (MKPs) have the capability to either recognize, bind, and dephosphorylate a singular class of MAP kinase or to modulate multiple MAPK pathways [163]. Genetic analyses and a BiFC assay were conducted to explore the role of MKP in regulating oxidative stress and pathogen defense responses, confirming that the MAPK phosphatase 2 (MKP2) and MPK3/MPK6 interaction occurs in both the cytoplasm and nucleus of Arabidopsis [164] (Figure 3E). In the scenario of fungal infection in Arabidopsis, the MKP2 and MPK3/MPK6 interaction can significantly reduce programmed cell death, referred to as the hypersensitive response (HR) [164], which is similar to the result of inhibiting MAPKs in plants [159]. Excessive accumulation of H2O2 can also trigger the HR in plants [165]. Evidence from Y2H and BiFC suggested that MPK31 in cotton Gossypium hirsutum (G. hirsutum) regulates the production of ROS and HR-like cell death through interaction with the H2O2-producing protein RBOHB [166]. Apart from the HR mediated by resistance proteins, plants can detect pathogens by surface-localized pattern recognition receptors (PRRs) via the recognition of carbohydrate-containing molecules such as fungal chitin, bacterial peptidoglycans, and extracellular ATP, which also activate multiple immune-related pathways, including the MAPK cascade [167]. It was revealed in rice (Oryza sativa L.) based on the BiFC assay that RLCK185 (receptor-like cytoplasmic kinase) regulates chitin-induced MAPK activation through interaction with MAPKKK11 and MAPKKK18 at the plasma membrane, and that MAPKKK18 interacts with MKK4, an upstream MAPKK component of MPK3/6, thus forming the MAPK cascade consisting of MAPKKK18–MKK4–MPK3/6 [168].
The MAPK cascade pathway is associated with the response to abiotic stress [169]. MAPKKK plays an important role as the largest gene family in the MAPK cascade [170]. To explore the physiological functions of MAPKKK in biotic and abiotic stress responses in the oilseed crop canola (Brassica napus L.), 15 interaction pairs between 28 MAPKKK proteins and 8 MAPKK proteins were identified via the Y2H assay and further validated through BiFC analysis [171]. The extensive interactions of MKK with MAP3K and MPK in G. hirsutum were revealed by Y2H and BiFC experiments, followed by VIGS (virus-induced gene silencing) assays to confirm the involvement of the MAP3K14-MKK11-MPK31 pathway in the drought stress response of cotton [172]. The subcellular localization and BiFC assays revealed that MAPK3 interacts with the proteins in the cold response pathway, ICE41, ICE87, and CBFIVd-D9 in the nucleus or in the plasma membrane of wheat (Triticum aestivum L.); this interaction is crucial for the negative regulation of plant cold tolerance, as MAPK3 mediates the phosphorylation and subsequent degradation of ICE and CBF proteins [173].
As an emerging biological technique, BiFC provides a powerful tool for deciphering cellular signaling pathways. The application of BiFC in cell signaling pathway research enables real-time and intuitive observation of PPIs, providing much convenience for pathway research. A large number of studies have successfully revealed the molecular mechanisms of various cellular signaling pathways using the BiFC technique. By using BiFC to gain a deeper understanding of molecular interactions, the regulatory mechanisms of signaling pathways can be revealed, providing key insights into cellular physiological processes, tissue development, and disease occurrence. In addition, this technology is expected to help identify new signaling pathway members and key regulatory factors, providing potential targets for the development of new drugs and treatment methods.
4. Protein—Protein Interactions Demonstrated by BiFC in Plant Hormone Signaling Pathways
In addition to being applied in the study of classic signaling pathways, BiFC has also been widely utilized in the investigation of plant hormones. The determination of the molecular interactions in the plant hormone pathways aids in elucidating the molecular mechanisms underlying plant growth, development, and stress responses, thereby laying the groundwork for the development of high-yield and stress-resistant plant varieties.
4.1. Ethylene Signaling Pathway
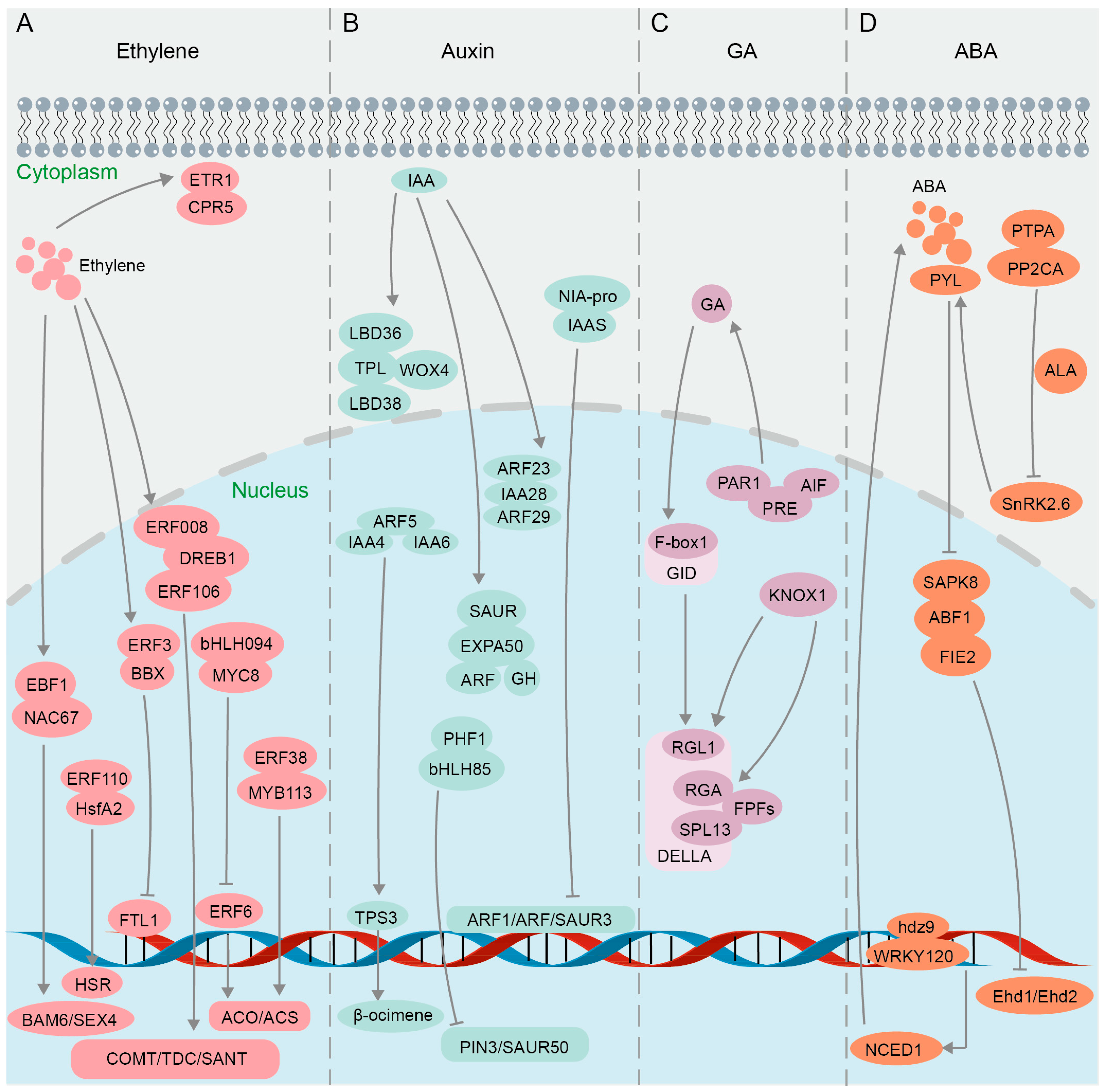
Ethylene accelerates fruit ripening, organ senescence and abscission, and promotes the differentiation of plant sexual organs [174]. The ethylene signaling pathway involves multiple families of transcription factors [175,176] (Figure 4A). Results of Y2H, GST pull-down, and BiFC assays confirmed the interaction between Ein3-binding F-box protein (MaEBF1) and MaNAC67-like protein, key components of the ethylene signaling pathway in Fenjiao banana (Musa ABB Pisang Awak), and their interaction further activates the promoters of starch degradation-related genes MaBAM6 and MaSEX4 (Figure 4A), thus facilitating fruit softening and ripening [177]. Yeast split-ubiquitin assays and BiFC studies indicated that Arabidopsis CPR5 (the constitutive expressor of pathogenesis-related genes 5) directly interacts with the ETR1 receptor in regulating ethylene signal transduction [178,179] (Figure 4A). BiFC and luciferase complementation imaging (LCI) assays confirmed that CPR5 in Melon (Cucumis melo L.) regulates the bisexual flower phenotype through interaction with ETR1 [180] (Figure 4A). The ethylene-responsive factor (ERF) belongs to the APETALA 2/ethylene response factor (AP2/ERF) superfamily and is a core component of the ethylene signaling pathway [181]. Multiple techniques including Y2H, BiFC, GST pull-down, and luciferase complementation imaging (LCI) were jointly utilized to discover that chrysanthemum CmERF3 inhibits the expression of flowering integrators (FTL1) by interacting with B-Box (BBX) family member CmBBX8 (Figure 4A), thereby maintaining the vegetative growth of Chrysanthemum morifolium and preventing premature flowering [182]. It was found in wheat (Triticum aestivum L.) through GST pull-down, Co-IP, Y2H, and BiFC assays that TabHLH094 (a basic helix–loop–helix transcription factor) and TaMYC8 (a negative regulator of cadmium-responsive ethylene signaling) form an interactive complex, reducing their ability to bind to the ERF6 promoter and thereby inhibiting the activities of 1-aminocyclopropane-1-carboxylate oxidase (ACO) and 1-aminocyclopropane-1-carboxylic acid synthase (ACS), two key rate-limiting enzymes in the process of plant ethylene biosynthesis [183] (Figure 4A). Results of Y2H, BiFC, and LCI assays showed that the protein ERF38 interacts with MYB113, enhancing the role of MYB113 on the promoters of ACS1 (Figure 4A), ultimately boosting transcriptional efficiency in eggplant (Solanum melongena L.) [184]. Ethylene can also regulate plant responses to abiotic stresses such as drought [185,186]. When plants experience heat stress, there is an elevation in the ethylene content present in their leaves [187]. Evidence from BiFC and Y2H assays of Lilium longiflorum showed that ERF110 interacts with the heat stress transcription factor (HsfA2) in the nucleus to regulate the expression of heat stress response (HSR) genes and improve heat tolerance [188] (Figure 4A). In the investigation of the functional mechanism of DREB1 in soybean (Glycine max L.), it was demonstrated through Y2H and BiFC experiments that two ERF-like transcription factors ERF008 and ERF106 interact with DREB1 (Figure 4A), thereby promoting the activation of drought-resistant genes COMT, TDC, and SANT [189]. In summary, the BiFC technique, either alone or in combination with other technologies, enables researchers to visualize PPIs and the subcellular localization of key components in the ethylene signaling pathway.
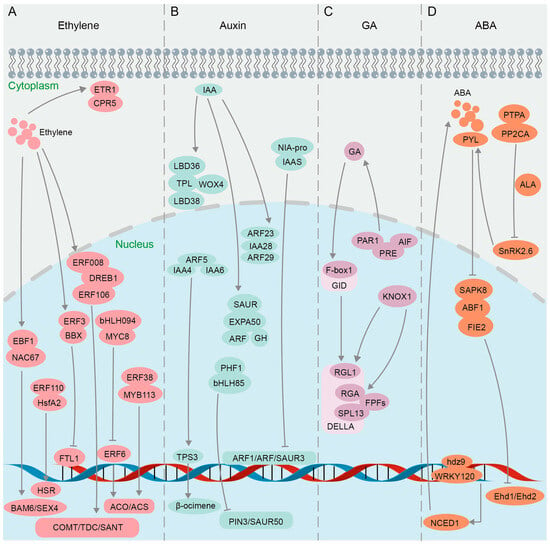
Figure 4.
Protein interactions in plant hormone pathways confirmed using BiFC. (A) Ethylene signaling pathway. (B) Auxin signaling pathway. (C) GA signaling pathway. (D) ABA signaling pathway.
4.2. Auxin Signaling Pathway
The growth hormone auxin mainly includes indole-3-acetic acid (IAA), indole-3-butyric acid (IBA), naphthoic acid (NAA), etc. Auxin plays a crucial role in various physiological processes in plants, such as promoting growth and development, enhancing stress tolerance, bolstering disease resistance, and improving resistance to herbicides [190,191]. The auxin pathway relies on two core regulatory factors, indole-3-acetic acid (IAA) protein and auxin response factors (ARFs) [192,193] (Figure 4B). It was confirmed by BiFC that ARF23 and ARF29 interact with IAA28 in the nucleus of Populus trichocarpa (P. trichocarpa) to participate in the IAA signal transduction process [194] (Figure 4B). IAA recruits TOPLESS (TPL) corepressors to inhibit the transcription of ARF auxin-responsive genes [195]. Y2H and BiFC assays demonstrated in Chrysanthemum morifolium that CmTPL1-1, a product of a Chrysanthemum TPL/TPR family gene, interacts with CmWOX4, CmLBD38, and CmLBD36 in modulating the auxin signaling pathway to regulate the development of root bud apical meristem and lateral organs [196] (Figure 4B). ARF5 interacts with odor-related auxin-responsive IAA4 and IAA6 in Hedychium coronarium to regulate the transcriptional activity of terpene synthase 3 (TPS3) in synthesizing volatile compounds related to floral fragrance β-ocimene, which was verified by Y2H and BiFC assays [197] (Figure 4B). When faced with pathogen infection, indole-3-acetic acid-amido synthetase (IAAS) induces the accumulation of conjugated auxin IAA-Asp, promoting the proliferation of pathogens [198]. It was confirmed through Y2H and BiFC that IAAS interacts with the Nia-pro protein encoded by Potato virus Y (PVY), inducing the expression of IAAS, while downregulating auxin-responsive genes, Auxin response factor 1 (ARF1), Auxin response factor 3 (ARF3), and small auxin upregulated RNA 3 (SAUR3) [199] (Figure 4B). Auxin enhances plant resistance to salt stress by inducing expansions and controlling cell wall plasticity [200]. It was proven through a BiFC assay that Chenopodium quinoa α-expansin 50 (CqEXPA50) interacts with the auxin pathway genes ARF, IAA, Gretchen Hagen 3 (GH3), and SAUR, resulting in the accumulation of photosynthetic pigments under salt stress [201] (Figure 4B). Y2H and BiFC evidence showed that the interaction between HLH85 and phosphate transporter chamber PHF1 in Sweet sorghum (Sorghum bicolor L.) disrupts the transport and accumulation of phosphorus (Pi) under salt stress and inhibits the expression of auxin pathway genes PIN3 and SAUR50 (Figure 4B), thereby reducing plant salt tolerance [202]. Collectively, by visualizing the interactions between auxin receptors and downstream signaling components, the BiFC technique has provided insights into the spatiotemporal dynamics and regulatory mechanisms of auxin signaling.
4.3. GA Signaling Pathway
Gibberellin (GA) signaling regulates various plant processes such as seed germination, stem elongation, root growth, flowering, and bud dormancy release in perennial woody plants [203,204,205,206,207,208]. The precise regulation of GA metabolism and signaling is crucial for plant growth and adaptation to the environment [209]. The GA signal is recognized by its nuclear receptor, GA INSENSITIVE DWARF1 (GID1), which initiates the GA signaling pathway by facilitating the interaction between GID1 and DELLA, a repressor of GA signaling [210]. GA regulates plant leaf bud dormancy and plant defense response through the GA-GID1-DELLA regulatory module, with DELLA proteins serving as key switches in GA signal transduction [211,212,213]. It was demonstrated through Y2H, BiFC, and GST pull-down assays that the C-terminal domain of PsF-box1 interacts with the DELLA member PsRGL1 in tobacco (Nicotiana benthamiana L.) leaves (Figure 4B), leading to the ubiquitination-dependent degradation of PsRGL1 and facilitating the release of tree peony bud dormancy [214]. The DELLA family members RGA1 and RGL1 interact with the hormone signaling regulator KNOX1 in Rape (Brassica campestris L.) (Figure 4B), as confirmed by Y2H and BiFC assays, facilitating bud differentiation and bolting via the GA pathway [215]. Flowering-promoting factors (FPFs) of Mango (Mangifera indica L.) were revealed through BiFC assays to interact with several DELLA proteins in positive regulation of both flowering promotion and root growth enhancement in response to GA treatment [216] (Figure 4B). The interaction between the auxin response factor and DELLA influences the GA and auxin signaling pathways, thereby modulating fruit development in tomato (Solanum lycopersicum L.) [217]. The overexpression of the atypical bHLH transcription factor SlPRE5 in tomatoes results in elevated levels of GA, and SlPRE5 interacts with bHLH proteins SlAIF1, SlAIF2, and SlPAR1 (Figure 4B), as demonstrated through Y2H and BiFC assays, to govern plant morphology and leaf chlorophyll accumulation, consequently impacting the process of photosynthesis [218]. Subsequently, it was elucidated through Y2H and BiFC assays that the interaction between an atypical basic bHLH transcription factor SlPRE3 and other bHLH proteins SlAIF1/SlAIF2/SlPAR1/SlIBH1 influences cell expansion and modulates lateral root growth associated with GA signaling [219] (Figure 4B). In conclusion, a growing body of research suggests that BiFC, either independently or in conjunction with other methodologies like Y2H and GST pull-down, offers a foundational comprehension of gene functionality in the GA signaling pathway. The BiFC assay will play a pivotal role in elucidating the complex interactions between GA and other hormonal signaling pathways, ultimately leading to the improvement of crop production and abiotic stress tolerance in plants.
4.4. ABA Signaling Pathway
The plant hormone abscisic acid (ABA) plays a pivotal role in regulating various physiological processes, particularly in response to abiotic stresses such as drought, salinity, and cold [220,221,222,223]. ABA and GA modulate plant growth and development through an antagonistic interplay [224]. The ABA signaling pathway is complex and involves numerous proteins that interact to transduce the hormonal signal into cellular responses (Figure 4D). In the context of the ABA signaling pathway, the BiFC assay has been used to decipher the interactions between key players. The bZIP transcription factor ABF1 was found to be phosphorylated by the SnRK2 (sucrose non-fermenting 1-related protein kinase 2) family member SAPK8 under exogenous ABA induction, and the interaction between ABF1 and SAPK8 was validated through Y2H, GST pull-down, BiFC, and kinase assays [225] (Figure 4B). Further investigation demonstrated that ABF1 interacts with the flowering negative regulatory factor FIE2 to recruit PRC2-mediated H3K27me3 modification to the target sites for inhibition, thereby delaying the flowering process of Oryza sativa [225]. In addition, SnRK2.6, a member of the SnRK2 family, activates the ABA signaling pathway and helps plants adapt to drought stress by controlling stomatal closure [226]. The results of Y2H, BiFC, and FLC (Firefly luciferase complementation imaging assay) experiments suggested that protein phosphatase type 2C group A (PP2AC) interacts with a photosynthetic phosphatase activator (PTPA) (Figure 4B), thereby exerting a negative regulatory effect on SnRK2.6 activity, ultimately resulting in stomatal opening in apple leaves [227]; their interaction was further verified by GST pull-down and MST (microscale thermophoresis) assays [228]. Furthermore, the results of Y2H and BiFC experiments indicated that the ABA receptor Pyrabastin Resistance 1-Like (PYL) interacts with PP2CA proteins in the presence of ABA signaling to establish a regulatory network that triggers stomatal closure in cotton Gossypium hirsutum L. during periods of drought [229]. Additional BiFC data also supported the notion that the interaction among PYL/PP2CA/SnRK2 proteins constitutes an ABA signal transduction module (Figure 4B), which plays a crucial role in mediating plant responses to drought stress [230,231]. Currently, there are reports indicating that members of the HD Zip transcription factor family, specifically ATHB-6 and HDZ5-6A, are involved in activating the ABA pathway in maize Zea mays L. and wheat Triticum aestivum L. to respond to drought stress [187,232]. Subsequently, it was confirmed through BiFC and GST pull-down experiments that Zmhdz9 interacts with ZmWRKY120 and ZmTCP9 (Figure 4B), promoting the expression of the key enzyme gene NCED1 in ABA biosynthesis, thereby enhancing drought resistance in Zea mays L. [233]. Taken together, the utilization of the BiFC assay in elucidating the ABA signaling pathway has been revolutionary, confirming established interactions and uncovering new components and controllers, thereby enhancing our understanding of this pivotal hormonal pathway.
In summary, the use of the BiFC technique to study protein–protein interactions in plant hormone pathways is of great significance for a deeper understanding of plant growth and development regulation, for revealing the molecular mechanisms by which plants respond to biotic and abiotic stresses, and also for developing new stress resistance technologies and optimizing agricultural production. In the future, the continuous application and progress of this technology will provide more scientific support and technological means for cultivating stress-tolerant varieties, promoting the development of precision agriculture and sustainable agriculture.
5. Optimization and Improvement of BiFC in Deciphering Protein–Protein Interactions
Despite the numerous advantages of the BiFC technique for visualizing PPIs, observing living cells in real-time, and analyzing subcellular localization, there are also limitations, including false-positive signals, limited spatial resolution, and weak fluorescence intensity, which necessitate ongoing optimization and improvement efforts (Figure 5A).
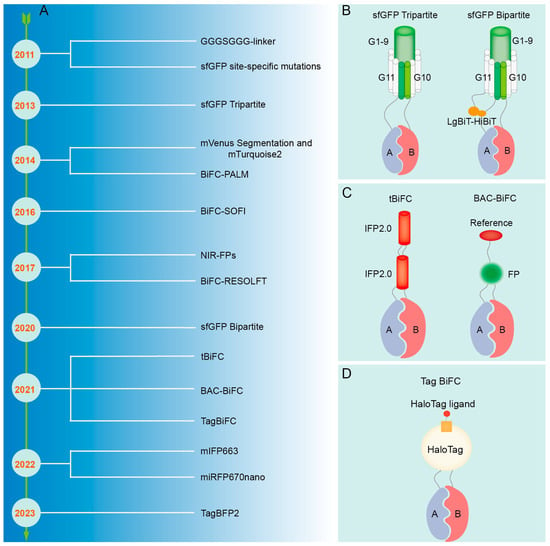
Figure 5.
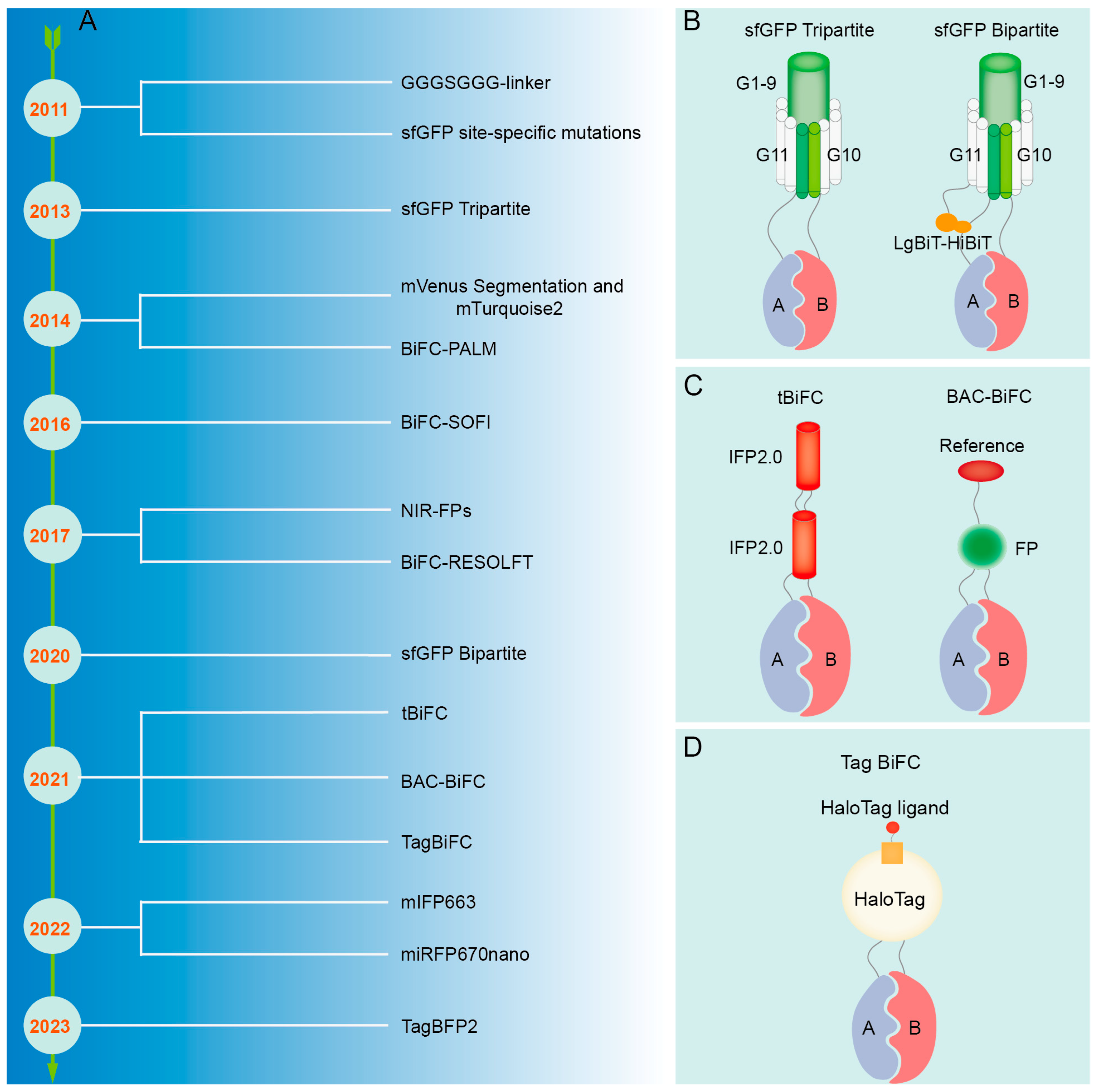
The refinement of the BiFC technique. (A) Improvement process of BiFC technique. (B) Schematic diagram of sfGFP. (C) Schematic diagram of tBiFC and BAC-BiFC. (D) Schematic diagram of Tag BiFC and sfGFP Bipartite.
5.1. Reduction of False-Positive Signal
False positives may arise due to nonspecific interactions of fluorescent fragments that self-assemble into a full fluorescent protein. A micro-tagging system, Tripartite Split-GFP, which is comprised of GFP10, GFP11, and detector GFP1-9 tags, minimizes protein interference and aggregation, and displays reduced background signals in mammalian cells, largely due to its concise labeling system [234] (Figure 5B). Mutations at certain amino acid residues located in the C-terminal fragment of Superfolder GFP (sfGFP), such as R219, D220, I229, and T230, have been demonstrated to hinder the self-assembly ability of sfGFP, resulting in a decrease in false-positive signals in negative controls [235] (Figure 5B). Additionally, segmentation at residue 210 of monomeric Venus (mVenus) can effectively decrease self-assembly, eliminate background signal, and precisely detect transformed cells in conjunction with the Golgi localization marker mTurquoise2 (mTq2) [80]. By incorporating an optical marker with one of the fragmented fluorescent proteins and utilizing it as a reference signal, the detection of non-specific PPIs can be prevented through the quantification of the fluorescence ratio between the BiFC signal and the reference signal; this method is referred to as BAC-BiFC (background assessable and correctable-bimolecular fluorescence complementation) [236] (Figure 5C). In BiFC experiments, the irreversible complementary binding of the N-terminal and C-terminal fragments of fluorescent proteins may result in artefactual readouts due to weak and transient interactions. Therefore, the inclusion of positive and negative controls is essential to assess false-positive signals and ensure the accuracy and reliability of experimental outcomes [61,64]. Cells co-transfected with the expression vectors pBiFC-bJunVN173 and pBiFC-bFosVC155 may serve as positive controls to assess the functionality of the experimental system. Conversely, cells transfected with plasmids solely encoding fluorescent protein fragments, plasmids encoding one target protein fused to fluorescent protein fragments, or plasmids encoding a non-interacting mutated partner can be utilized as negative controls.
5.2. Improvement of Spatial Resolution
The conventional BiFC resolution is limited by light diffraction, making it inappropriate for detecting PPI at the nanoscale. When utilized in conjunction with Photoactivated Localization Microscopy (PALM) [237] and Stochastic Optical Fluctuation Imaging (SOFI) [238], the enhanced BiFC technique can be employed for the precise localization of PPI at the nanoscale level. Nevertheless, BiFC-PALM is limited to observing interacting proteins in fixed cells [237,239], whereas BiFC-SOFI necessitates imaging analysis for obtaining super-resolution images, rendering it unsuitable for high-resolution real-time PPI studies in live cells [238]. Real-time monitoring of PPI within living cells was achieved at the nanoscale resolution using Reversible Saturable Optical Fluorescence Transition (RESOLFT) nanoscopy [240]. When traditional Fluorescent Protein Indicators are transformed into photo-transformable Fluorescent Protein-based Indicators (ptFP), such as PAmCherry1 [237], mEos3.2 [239], mIrisFP [241], and rsEGFP2 [240], the identification of their cleavage sites can be determined by analyzing their individual protein structures. This approach involves the integration of super-resolution microscopy (SRM) techniques (BiFC-PALM, BiFC-SOFI, and BiFC-RESOLFT) to enable super-resolution imaging of protein interactions [242]. Furthermore, the proper manipulation of experimental materials, such as the deliberate creation of gaps between the lower epidermis and muscle layer tissues of plant leaves, has been shown to mitigate the potential interference of the muscle layer and upper epidermis on imaging quality, ultimately enhancing resolution [243].
5.3. Enhancement of Fluorescence Signal
Traditional BiFC methods are hindered by low signal intensity and instability, thereby restricting their capacity to identify interactions between proteins with weak affinity. The GGGSGGG-linker sequence was employed for the purpose of connecting mRFP fragments with target proteins, enhancing the flexibility of the fusion protein interaction process; this optimization has been shown to have a notable impact on signal strength [244] (Figure 5B). The improved Tripartite sfGFP can significantly enhance the fluorescence signal by capturing the interacting protein complex with the anti-GFP(1-9) nanobody (VHHr) [245]. The incorporation of Luciferase Bioluminescence Technology-High Bioluminescence Tag (LgBiT-HiBiT) onto the GFP1-9 and GFP11 fragments results in the conversion of the sfGFP Tripartite system into a novel Bipartite system, thereby substantially improving the interaction signal and signal-to-noise ratio of BiFC detection [246] (Figure 5B). The tandem near-infrared BiFC system (tBiFC) was developed by linking two fragments of IFP2.0 (Improved monomeric near-infrared phytochrome 2.0) [76] (Figure 5C) or utilizing miRFP670nano as the signal module [75], enhancing the intensity and sensitivity of the BiFC signal while exhibiting superior optical stability. Organic dyes exhibit superior brightness and light stability compared to fluorescent proteins and can be conjugated to target proteins through self-labeling [247,248]. Consequently, the Tagged Bimolecular Fluorescence Complementation (TagBiFC) system, which utilizes the Haloalkane Dehalogenase Tag (HaloTag), offers enhanced signal intensity for visualizing PPIs in live cell imaging [249] (Figure 5D). Although monomeric near-infrared (NIR) fluorescent proteins (FPs) have a longer wavelength compared to infrared FP (IFP) and possess attributes such as low cytotoxicity and suitability for deep penetration imaging, they exhibit relatively low brightness when excited by conventional lasers [250]. The monomeric IFP 663m (mIFP663) was utilized as a fluorophore to optimize excitation at 633 nm, exhibiting superior brightness, stability, and compatibility with subcellular localization in the PPI analysis of viable cells [251]. Moreover, tagged blue fluorescent protein 2 (TagBFP2) exhibits superior brightness and a superior signal-to-noise ratio in BiFC detection when compared to conventional fluorescent proteins, enabling the acquisition of high-quality images without the requirement of costly high-end equipment [15].
6. Concluding Remarks and Future Perspectives
The cell signaling pathway is an important process of information transfer within and between cells, which regulates cell growth, development, and function. Protein–protein interactions are one of the core mechanisms by which cell signaling pathways function. BiFC provides a way to visualize and quantify protein interactions for cell biology research. The application of BiFC in cell signal transduction research is summarized as follows: (1) The detection of protein interactions: by directly observing fluorescence signals, the interaction between proteins can be detected in real-time and dynamically, so the BiFC assay allows us to understand the regulatory mechanisms of cell signal transduction pathways. (2) Uncovering the composition of protein complexes: by combining with other protein-labeling techniques, such as immunolabeling or genetic labeling, BiFC can be used to identify the composition of protein complexes involved in specific signal transduction events. (3) Revealing the subcellular localization of protein interactions: luminescence of fluorescent proteins can provide subcellular localization information of protein interactions, which is critical for understanding the spatial and temporal regulation of specific signal transduction events within cells. (4) Drug screening and functional research: BiFC can be used to screen compounds or small molecules that can affect specific protein interactions, further providing candidate molecules for drug discovery. In addition, BiFC can also be used to study the effects of protein interactions on cell functions, such as proliferation, differentiation, and apoptosis.
The BiFC technique has a broad application prospect in the research of cell signaling pathways, and its future development trend is concentrated in the following aspects: (1) Improving signal intensity and sensitivity: enhancing the fluorescence signal intensity and sensitivity of BiFC will enable the detection and quantitative analysis of proteins with low expression levels or weak interactions, providing more accurate insights into protein interaction events. (2) Combining with multi-channel and multi-labeling techniques: by introducing fluorescent proteins of different colors or using fluorescent dyes for multi-channel labeling, multiple interaction events within the same cell can be detected simultaneously, enabling a more comprehensive understanding of the complexity of gene regulatory networks. (3) High-resolution imaging and 3D visualization: With the advancement of microscopy technology, more attention will be paid to high-resolution imaging and 3D visualization. The combination of super-resolution microscopy and imaging techniques will allow for the observation and quantitative analysis of dynamic changes in protein interactions at the subcellular level [252]. (4) Combining with high-throughput omics technology: by integrating large-scale protein interaction data and expression data, a more accurate and complete gene regulatory network model is constructed. (5) Utilizing molecular-docking and machine-learning methodologies for the prediction of PPI networks [253] and subsequently corroborating these predictions through the application of BiFC to validate the interactions of the relevant proteins. For example, AlphaFold3 exhibits exceptional capability in accurately predicting the high-precision structural interactions of various biological molecules [254].
The current BiFC technique, including in vivo applications, necessitates the use of plasmids for the expression of exogenous proteins to detect protein interactions. Thus, the challenge of observing endogenous protein interactions in vivo remains a significant issue to be addressed. Miyakura et al. [155] fused the N-terminal of Kusabira-Green (mKG) with the decoy protein and then used PiggyBac transposon to insert the sequence of the remaining mKG into the genome, randomly fusing endogenous genes and combining RACE experiments to achieve the screening of endogenous interacting proteins. This method provides a new way to observe the interaction of endogenous proteins, but the expression of the decoy protein requires the introduction of foreign plasmids. It may be possible to insert the expression sequence of fluorescent protein fragments into the genome at a fixed point, but the selection of insertion sites, the amount of endogenous protein expression, and the intensity of fluorescent protein need to be verified. Combining BiFC with other technologies, such as CRISPR-Cas9 gene editing [255,256], live-cell imaging, and ultra-high-resolution microscopy imaging [252], will further improve our understanding of protein interactions in cell signaling pathways.
Author Contributions
H.R.: Writing—original draft, Visualization, Investigation, Conceptualization. Q.O.: Writing—review and editing. Q.P.: Writing—review and editing. Y.L.: Writing—review and editing. X.Y.: Writing—review and editing. Y.H.: Writing—review and editing. S.L.: Writing—original draft, Review and editing, Visualization, Supervision, Funding acquisition, Conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32172797), Chongqing Science and Technology Plan Project (CSTB2022NSCQ-MSX0491), Chongqing Graduate Student Research Innovation Project (CYB21132; CYB240144), and Fundamental Research Funds for the Central Universities (SWU-XJPY202309).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No data were generated for the research described in this article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Iqbal, M.J.; Kabeer, A.; Abbas, Z.; Siddiqui, H.A.; Calina, D.; Sharifi-Rad, J.; Cho, W.C. Interplay of oxidative stress, cellular communication and signaling pathways in cancer. Cell Commun. Signal. 2024, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Kawano, T.; Murata, M.; Toita, R. Vascular calcification and cellular signaling pathways as potential therapeutic targets. Life Sci. 2024, 336, 122309. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Berry, G.J.; Liang, D.H.; Goronzy, J.J.; Weyand, C.M. Cellular Signaling Pathways in Medium and Large Vessel Vasculitis. Front. Immunol. 2020, 11, 587089. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, M.; Safari, F.; Sobhani, N.; Alavi, M.; Roudi, R. Regulation of Cellular-Signaling Pathways by Mammalian Proteins Containing Bacterial EPIYA or EPIYA-Like Motifs Predicted to be Phosphorylated. DNA Cell Biol. 2024, 43, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Hashemi Karoii, D.; Azizi, H. A review of protein-protein interaction and signaling pathway of Vimentin in cell regulation, morphology and cell differentiation in normal cells. J. Recept. Signal Transduct. Res. 2022, 42, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Cusick, M.E.; Klitgord, N.; Vidal, M.; Hill, D.E. Interactome: Gateway into systems biology. Hum. Mol. Genet. 2005, 14, R171–R181. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Bhawal, R.; Yin, Z.; Thannhauser, T.W.; Zhang, S. Recent advances in proteomics and metabolomics in plants. Mol. Hortic. 2022, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Struk, S.; Jacobs, A.; Sanchez Martin-Fontecha, E.; Gevaert, K.; Cubas, P.; Goormachtig, S. Exploring the protein-protein interaction landscape in plants. Plant Cell Environ. 2019, 42, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: Characteristics of protein fragment complementation. Chem. Soc. Rev. 2009, 38, 2876–2886. [Google Scholar] [CrossRef]
- Avilov, S.V.; Aleksandrova, N. Fluorescence protein complementation in microscopy: Applications beyond detecting bi-molecular interactions. Methods Appl. Fluoresc. 2018, 7, 012001. [Google Scholar] [CrossRef]
- Miller, K.E.; Kim, Y.; Huh, W.K.; Park, H.O. Bimolecular Fluorescence Complementation (BiFC) Analysis: Advances and Recent Applications for Genome-Wide Interaction Studies. J. Mol. Biol. 2015, 427, 2039–2055. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S. Bimolecular fluorescence complementation: Illuminating cellular protein interactions. Curr. Mol. Med. 2011, 11, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Purkait, D.; Ilyas, M.; Atmakuri, K. Protein-Protein Interactions: Bimolecular Fluorescence Complementation and Cytology Two Hybrid. Methods Mol. Biol. 2024, 2715, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, M.; Liu, T.; Qin, K.; Fernie, A.R. Investigating the dynamics of protein-protein interactions in plants. Plant J. 2023, 114, 965–983. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Gao, X.; Zhang, W.; Chen, B.; Wang, M.; Liao, K.; Wang, Z.; Ren, L.; Zhai, Y.; Qiu, Y.; et al. Novel Bimolecular Fluorescence Complementation (BiFC) Assay for Visualization of the Protein-Protein Interactions and Cellular Protein Complex Localizations. Mol. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, J. Imaging Spatial Reorganization of a MAPK Signaling Pathway Using the Tobacco Transient Expression System. J. Vis. Exp. 2016, 109, e53790. [Google Scholar] [CrossRef]
- Riese, F.; Grinschgl, S.; Gersbacher, M.T.; Russi, N.; Hock, C.; Nitsch, R.M.; Konietzko, U. Visualization and quantification of APP intracellular domain-mediated nuclear signaling by bimolecular fluorescence complementation. PLoS ONE 2013, 8, e76094. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.Y.; Abere, B.; Shi, H.; Toland, S.; Smithgall, T.E.; Moore, P.S.; Chang, Y. Membrane-bound Merkel cell polyomavirus middle T protein constitutively activates PLCgamma1 signaling through Src-family kinases. Proc. Natl. Acad. Sci. USA 2023, 120, e2316467120. [Google Scholar] [CrossRef]
- Ma, H.; Zou, F.; Li, D.; Wan, Y.; Zhang, Y.; Zhao, Z.; Wang, X.; Gao, H. Transcription Factor MdbHLH093 Enhances Powdery Mildew Resistance by Promoting Salicylic Acid Signaling and Hydrogen Peroxide Accumulation. Int. J. Mol. Sci. 2023, 24, 9390. [Google Scholar] [CrossRef]
- Ding, Y.; Su, S.; Tang, W.; Zhang, X.; Chen, S.; Zhu, G.; Liang, J.; Wei, W.; Guo, Y.; Liu, L.; et al. Enrichment of the β-catenin-TCF complex at the S and G2 phases ensures cell survival and cell cycle progression. J. Cell Sci. 2014, 127 Pt 22, 4833–4845. [Google Scholar] [CrossRef]
- Sung, M.K.; Huh, W.K. In vivo quantification of protein-protein interactions in Saccharomyces cerevisiae using bimolecular fluorescence complementation assay. J. Microbiol. Methods 2010, 83, 194–201. [Google Scholar] [CrossRef]
- Yu, W.; Xu, X.; Jin, K.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Genetically encoded biosensors for microbial synthetic biology: From conceptual frameworks to practical applications. Biotechnol. Adv. 2023, 62, 108077. [Google Scholar] [CrossRef]
- Cabantous, S.; Waldo, G.S. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods 2006, 3, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Listwan, P.; Terwilliger, T.C.; Waldo, G.S. Automated, high-throughput platform for protein solubility screening using a split-GFP system. J. Struct. Funct. Genomics 2009, 10, 47–55. [Google Scholar] [CrossRef]
- Kent, K.P.; Boxer, S.G. Light-activated reassembly of split green fluorescent protein. J. Am. Chem. Soc. 2011, 133, 4046–4052. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.D.; Kerppola, T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003, 21, 539–545. [Google Scholar] [CrossRef]
- Beinecke, F.A.; Grundmann, L.; Wiedmann, D.R.; Schmidt, F.J.; Caesar, A.S.; Zimmermann, M.; Lahme, M.; Twyman, R.M.; Prufer, D.; Noll, G.A. The FT/FD-dependent initiation of flowering under long-day conditions in the day-neutral species Nicotiana tabacum originates from the facultative short-day ancestor Nicotiana tomentosiformis. Plant J. 2018, 96, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Marczak, M.; Ciesla, A.; Janicki, M.; Kasprowicz-Maluski, A.; Kubiak, P.; Ludwikow, A. Protein Phosphatases Type 2C Group A Interact with and Regulate the Stability of ACC Synthase 7 in Arabidopsis. Cells 2020, 9, 978. [Google Scholar] [CrossRef] [PubMed]
- Boruc, J.; Inze, D.; Russinova, E. A high-throughput bimolecular fluorescence complementation protein-protein interaction screen identifies functional Arabidopsis CDKA/B-CYCD4/5 complexes. Plant Signal. Behav. 2010, 5, 1276–1281. [Google Scholar] [CrossRef]
- Wang, L.; Carnegie, G.K. Flow cytometric analysis of bimolecular fluorescence complementation: A high throughput quantitative method to study protein-protein interaction. J. Vis. Exp. 2013, 78, e50529. [Google Scholar] [CrossRef]
- Nishimura, K.; Ishikawa, S.; Matsunami, E.; Yamauchi, J.; Homma, K.; Faulkner, C.; Oparka, K.; Jisaka, M.; Nagaya, T.; Yokota, K.; et al. New Gateway-compatible vectors for a high-throughput protein-protein interaction analysis by a bimolecular fluorescence complementation (BiFC) assay in plants and their application to a plant clathrin structure analysis. Biosci. Biotechnol. Biochem. 2015, 79, 1995–2006. [Google Scholar] [CrossRef] [PubMed]
- Lepur, A.; Kovacevic, L.; Beluzic, R.; Vugrek, O. Combining Unique Multiplex Gateway Cloning and Bimolecular Fluorescence Complementation (BiFC) for High-Throughput Screening of Protein-Protein Interactions. J. Biomol. Screen. 2016, 21, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zhu, D.; Zhang, Z.; Wang, W.; Fan, J.; Men, D.; Deng, J.; Wei, H.; Zhang, X.E.; Cui, Z. Imaging of mRNA-protein interactions in live cells using novel mCherry trimolecular fluorescence complementation systems. PLoS ONE 2013, 8, e80851. [Google Scholar] [CrossRef]
- Han, Y.; Wang, S.; Zhang, Z.; Ma, X.; Li, W.; Zhang, X.; Deng, J.; Wei, H.; Li, Z.; Zhang, X.E.; et al. In vivo imaging of protein-protein and RNA-protein interactions using novel far-red fluorescence complementation systems. Nucleic Acids Res. 2014, 42, e103. [Google Scholar] [CrossRef]
- Chen, M.; Sui, T.; Yang, L.; Qian, Y.; Liu, Z.; Liu, Y.; Wang, G.; Lai, L.; Li, Z. Live imaging of RNA and RNA splicing in mammalian cells via the dcas13a-SunTag-BiFC system. Biosens. Bioelectron. 2022, 204, 114074. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, H.; Wang, S.; Ding, M.; An, H.; Hou, Y.; Yang, X.; Wei, W.; Sun, Y.; Tang, C. Live visualization of genomic loci with BiFC-TALE. Sci. Rep. 2017, 7, 40192. [Google Scholar] [CrossRef]
- Zhu, J.; He, X.; Bernard, D.; Shen, J.; Su, Y.; Wolek, A.; Issacs, B.; Mishra, N.; Tian, X.; Garmendia, A.; et al. Identification of New Compounds against PRRSV Infection by Directly Targeting CD163. J. Virol. 2023, 97, e0005423. [Google Scholar] [CrossRef]
- Hinz, S.; Navarro, G.; Borroto-Escuela, D.; Seibt, B.F.; Ammon, Y.C.; de Filippo, E.; Danish, A.; Lacher, S.K.; Cervinkova, B.; Rafehi, M.; et al. Adenosine A2A receptor ligand recognition and signaling is blocked by A2B receptors. Oncotarget 2018, 9, 13593–13611. [Google Scholar] [CrossRef]
- Zych, C.; Domling, A.; Ayyavoo, V. Development of a robust cell-based high-throughput screening assay to identify targets of HIV-1 viral protein R dimerization. Drug Des. Devel. Ther. 2013, 7, 403–412. [Google Scholar] [CrossRef]
- Kaczor, A.A.; Selent, J. Oligomerization of G protein-coupled receptors: Biochemical and biophysical methods. Curr. Med. Chem. 2011, 18, 4606–4634. [Google Scholar] [CrossRef]
- Poe, J.A.; Vollmer, L.; Vogt, A.; Smithgall, T.E. Development and validation of a high-content bimolecular fluorescence complementation assay for small-molecule inhibitors of HIV-1 Nef dimerization. J. Biomol. Screen. 2014, 19, 556–565. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, R.; Leu, N.A.; Zhang, L.; Ibragmova, I.; Schultz, D.C.; Wang, P.J. A cell-based high-content screen identifies isocotoin as a small molecule inhibitor of the meiosis-specific MEIOB-SPATA22 complexdagger. Biol. Reprod. 2020, 103, 333–342. [Google Scholar] [CrossRef]
- Hwang, H.Y.; Shim, J.S.; Kim, D.; Kwon, H.J. Antidepressant drug sertraline modulates AMPK-MTOR signaling-mediated autophagy via targeting mitochondrial VDAC1 protein. Autophagy 2021, 17, 2783–2799. [Google Scholar] [CrossRef] [PubMed]
- Bellon-Echeverria, I.; Carralot, J.P.; Del Rosario, A.A.; Kueng, S.; Mauser, H.; Schmid, G.; Thoma, R.; Berger, I. MultiBacMam Bimolecular Fluorescence Complementation (BiFC) tool-kit identifies new small-molecule inhibitors of the CDK5-p25 protein-protein interaction (PPI). Sci. Rep. 2018, 8, 5083. [Google Scholar] [CrossRef]
- Doyle, T.B.; Muntean, B.S.; Ejendal, K.F.; Hayes, M.P.; Soto-Velasquez, M.; Martemyanov, K.A.; Dessauer, C.W.; Hu, C.D.; Watts, V.J. Identification of Novel Adenylyl Cyclase 5 (AC5) Signaling Networks in D1 and D2 Medium Spiny Neurons using Bimolecular Fluorescence Complementation Screening. Cells 2019, 8, 1468. [Google Scholar] [CrossRef] [PubMed]
- Lashuel, H.A. Alpha-Synuclein oligomerization and aggregation: All models are useful but only if we know what they model. J. Neurochem. 2021, 157, 891–898. [Google Scholar] [CrossRef]
- Thakur, N.; Conceicao, C.; Isaacs, A.; Human, S.; Modhiran, N.; McLean, R.K.; Pedrera, M.; Tan, T.K.; Rijal, P.; Townsend, A.; et al. Micro-fusion inhibition tests: Quantifying antibody neutralization of virus-mediated cell-cell fusion. J. Gen. Virol. 2021, 102, 001506. [Google Scholar] [CrossRef] [PubMed]
- Eckermann, K.; Kugler, S.; Bahr, M. Dimerization propensities of Synucleins are not predictive for Synuclein aggregation. Biochim. Biophys. Acta 2015, 1852, 1658–1664. [Google Scholar] [CrossRef][Green Version]
- Ji, Y.; Jiang, J.; Huang, L.; Feng, W.; Zhang, Z.; Jin, L.; Xing, X. Sperm-associated antigen 4 (SPAG4) as a new cancer marker interacts with Nesprin3 to regulate cell migration in lung carcinoma. Oncol. Rep. 2018, 40, 783–792. [Google Scholar] [CrossRef]
- Jahreis, K.; Bruge, A.; Borsdorf, S.; Muller, F.E.; Sun, W.; Jia, S.; Kang, D.M.; Boesen, N.; Shin, S.; Lim, S.; et al. Amisulpride as a potential disease-modifying drug in the treatment of tauopathies. Alzheimer’s Dement. 2023, 19, 5482–5497. [Google Scholar] [CrossRef]
- Yu, X.; Munoz-Sagredo, L.; Streule, K.; Muschong, P.; Bayer, E.; Walter, R.J.; Gutjahr, J.C.; Greil, R.; Concha, M.L.; Muller-Tidow, C.; et al. CD44 loss of function sensitizes AML cells to the BCL-2 inhibitor venetoclax by decreasing CXCL12-driven survival cues. Blood 2021, 138, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, S.J.; Jo, D.S.; Park, N.Y.; Kim, J.B.; Bae, J.E.; Jo, Y.K.; Hwang, J.J.; Lee, J.A.; Jo, D.G.; et al. Down-regulated TMED10 in Alzheimer disease induces autophagy via ATG4B activation. Autophagy 2019, 15, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Fuxe, K.; Marcellino, D.; Borroto-Escuela, D.O.; Guescini, M.; Fernandez-Duenas, V.; Tanganelli, S.; Rivera, A.; Ciruela, F.; Agnati, L.F. Adenosine-dopamine interactions in the pathophysiology and treatment of CNS disorders. CNS Neurosci. Ther. 2010, 16, e18–e42. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qu, X.; Guan, C.; Luo, N.; Chen, H.; Li, A.; Zhuang, H.; Yang, J.; Diao, H.; Zeng, S.; et al. Mitochondrial micropeptide MOXI promotes fibrotic gene transcription by translocation to the nucleus and bridging N-acetyltransferase 14 with transcription factor c-Jun. Kidney Int. 2023, 103, 886–902. [Google Scholar] [CrossRef] [PubMed]
- Kodama, Y.; Hu, C.D. Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 2012, 53, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006, 1, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 2008, 37, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, N.; Varshavsky, A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA 1994, 91, 10340–10344. [Google Scholar] [CrossRef] [PubMed]
- Dunnwald, M.; Varshavsky, A.; Johnsson, N. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol. Biol. Cell 1999, 10, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Chadwick, C.S.; Nairn, R.C. Fluorescent protein tracers: The unreacted fluorescent material in fluorescein conjugates and studies of conjugates with other green fluorochromes. Immunology 1960, 3, 363–370. [Google Scholar]
- Hu, C.D.; Chinenov, Y.; Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Prasher, D.C.; Eckenrode, V.K.; Ward, W.W.; Prendergast, F.G.; Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 1992, 111, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, I.; Hamilton, A.D.; Regan, L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J. Am. Chem. Soc. 2000, 122, 5658–5659. [Google Scholar] [CrossRef]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 2006, 40, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Shyu, Y.J.; Suarez, C.D.; Hu, C.D. Visualization of AP-1 NF-kappaB ternary complexes in living cells by using a BiFC-based FRET. Proc. Natl. Acad. Sci. USA 2008, 105, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Jach, G.; Pesch, M.; Richter, K.; Frings, S.; Uhrig, J.F. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat. Methods 2006, 3, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Y.; Cui, Z.Q.; Wei, H.P.; Zhang, Z.P.; Zhou, Y.F.; Wang, Y.P.; Zhang, X.E. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem. Biophys. Res. Commun. 2008, 367, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Zhang, Z.; Zheng, Y.; Yang, J.; Qin, L.; Lu, J.; Huang, Z.L.; Zeng, S.; Luo, Q. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens. Bioelectron. 2009, 25, 234–239. [Google Scholar] [CrossRef]
- Wang, S.; Ding, M.; Xue, B.; Hou, Y.; Sun, Y. Spying on protein interactions in living cells with reconstituted scarlet light. Analyst 2018, 143, 5161–5169. [Google Scholar] [CrossRef]
- Fujii, Y.; Yoshimura, A.; Kodama, Y. A novel orange-colored bimolecular fluorescence complementation (BiFC) assay using monomeric Kusabira-Orange protein. Biotechniques 2018, 64, 153–161. [Google Scholar] [CrossRef]
- Peter, S.; Oven-Krockhaus, S.Z.; Veerabagu, M.; Rodado, V.M.; Berendzen, K.W.; Meixner, A.J.; Harter, K.; Schleifenbaum, F.E. Chimeric Autofluorescent Proteins as Photophysical Model System for Multicolor Bimolecular Fluorescence Complementation. J. Phys. Chem. B 2017, 121, 2407–2419. [Google Scholar] [CrossRef]
- Fankhauser, C. The phytochromes, a family of red/far-red absorbing photoreceptors. J. Biol. Chem. 2001, 276, 11453–11456. [Google Scholar] [CrossRef]
- Chen, M.; Li, W.; Zhang, Z.; Liu, S.; Zhang, X.; Zhang, X.E.; Cui, Z. Novel near-infrared BiFC systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions. Biomaterials 2015, 48, 97–107. [Google Scholar] [CrossRef]
- Tchekanda, E.; Sivanesan, D.; Michnick, S.W. An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat. Methods 2014, 11, 641–644. [Google Scholar] [CrossRef]
- Chen, M.; Yan, C.; Zheng, L.; Zhang, X.E. The smallest near-infrared fluorescence complementation system for imaging protein-protein and RNA-protein interactions. Chem. Sci. 2022, 13, 1119–1129. [Google Scholar] [CrossRef]
- Chen, M.; Yan, C.; Ma, Y.; Zhang, X.E. A tandem near-infrared fluorescence complementation system with enhanced fluorescence for imaging protein-protein interactions in vivo. Biomaterials 2021, 268, 120544. [Google Scholar] [CrossRef]
- Lee, L.Y.; Fang, M.J.; Kuang, L.Y.; Gelvin, S.B. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 2008, 4, 24. [Google Scholar] [CrossRef]
- Gehl, C.; Waadt, R.; Kudla, J.; Mendel, R.R.; Hansch, R. New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2009, 2, 1051–1058. [Google Scholar] [CrossRef]
- Grefen, C.; Blatt, M.R. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 2012, 53, 311–314. [Google Scholar] [CrossRef]
- Gookin, T.E.; Assmann, S.M. Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 2014, 80, 553–567. [Google Scholar] [CrossRef]
- Luo, Y.; Qiu, Y.; Na, R.; Meerja, F.; Lu, Q.S.; Yang, C.; Tian, L. A Golden Gate and Gateway double-compatible vector system for high throughput functional analysis of genes. Plant Sci. 2018, 271, 117–126. [Google Scholar] [CrossRef]
- Han, J.; Ma, K.; Li, H.; Su, J.; Zhou, L.; Tang, J.; Zhang, S.; Hou, Y.; Chen, L.; Liu, Y.G.; et al. All-in-one: A robust fluorescent fusion protein vector toolbox for protein localization and BiFC analyses in plants. Plant Biotechnol. J. 2022, 20, 1098–1109. [Google Scholar] [CrossRef]
- Ren, R.; Gao, J.; Lu, C.; Wei, Y.; Jin, J.; Wong, S.M.; Zhu, G.; Yang, F. Highly Efficient Protoplast Isolation and Transient Expression System for Functional Characterization of Flowering Related Genes in Cymbidium Orchids. Int. J. Mol. Sci. 2020, 21, 2264. [Google Scholar] [CrossRef]
- Velay, F.; Soula, M.; Mehrez, M.; Belbachir, C.; D’Alessandro, S.; Laloi, C.; Crete, P.; Field, B. MoBiFC: Development of a modular bimolecular fluorescence complementation toolkit for the analysis of chloroplast protein-protein interactions. Plant Methods 2022, 18, 69. [Google Scholar] [CrossRef]
- Prinz, W.A.; Toulmay, A.; Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020, 21, 7–24. [Google Scholar] [CrossRef]
- Zhou, M.; Kong, B.; Zhang, X.; Xiao, K.; Lu, J.; Li, W.; Li, M.; Li, Z.; Ji, W.; Hou, J.; et al. A proximity labeling strategy enables proteomic analysis of inter-organelle membrane contacts. iScience 2023, 26, 107159. [Google Scholar] [CrossRef]
- Maika, J.E.; Kramer, B.; Strotmann, V.I.; Wellmer, F.; Weidtkamp-Peters, S.; Stahl, Y.; Simon, R. One pattern analysis (OPA) for the quantitative determination of protein interactions in plant cells. Plant Methods 2023, 19, 73. [Google Scholar] [CrossRef]
- Lee, O.H.; Kim, H.; He, Q.; Baek, H.J.; Yang, D.; Chen, L.Y.; Liang, J.; Chae, H.K.; Safari, A.; Liu, D.; et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell. Proteom. MCP 2011, 10, S1–S11. [Google Scholar] [CrossRef]
- Bischof, J.; Duffraisse, M.; Furger, E.; Ajuria, L.; Giraud, G.; Vanderperre, S.; Paul, R.; Bjorklund, M.; Ahr, D.; Ahmed, A.W.; et al. Generation of a versatile BiFC ORFeome library for analyzing protein-protein interactions in live Drosophila. Elife 2018, 7, e38853. [Google Scholar] [CrossRef]
- Jia, Y.; Reboulet, J.; Gillet, B.; Hughes, S.; Forcet, C.; Tribollet, V.; Hajj Sleiman, N.; Kundlacz, C.; Vanacker, J.M.; Bleicher, F.; et al. A Live Cell Protein Complementation Assay for ORFeome-Wide Probing of Human HOX Interactomes. Cells 2023, 12, 200. [Google Scholar] [CrossRef]
- Miyake, T.; McDermott, J.C. Functional analysis of protein interactions using coupled bi-fluorescence complementation/GFP nanobody techniques. Nucleic Acids Res. 2024, gkae548. [Google Scholar] [CrossRef]
- Gomes, A.P.; Blenis, J. A nexus for cellular homeostasis: The interplay between metabolic and signal transduction pathways. Curr. Opin. Biotechnol. 2015, 34, 110–117. [Google Scholar] [CrossRef]
- Shimobayashi, M.; Hall, M.N. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 2014, 15, 155–162. [Google Scholar] [CrossRef]
- Efeyan, A.; Sabatini, D.M. mTOR and cancer: Many loops in one pathway. Curr. Opin. Cell Biol. 2010, 22, 169–176. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Yoon, M.S.; Rosenberger, C.L.; Wu, C.; Truong, N.; Sweedler, J.V.; Chen, J. Rapid mitogenic regulation of the mTORC1 inhibitor, DEPTOR, by phosphatidic acid. Mol. Cell 2015, 58, 549–556. [Google Scholar] [CrossRef]
- Guan, B.; Jiang, Y.T.; Lin, D.L.; Lin, W.H.; Xue, H.W. Phosphatidic acid suppresses autophagy through competitive inhibition by binding GAPC (glyceraldehyde-3-phosphate dehydrogenase) and PGK (phosphoglycerate kinase) proteins. Autophagy 2022, 18, 2656–2670. [Google Scholar] [CrossRef]
- Zhuang, X.; Chung, K.P.; Luo, M.; Jiang, L. Autophagosome Biogenesis and the Endoplasmic Reticulum: A Plant Perspective. Trends Plant Sci. 2018, 23, 677–692. [Google Scholar] [CrossRef]
- Michaeli, S.; Clavel, M.; Lechner, E.; Viotti, C.; Wu, J.; Dubois, M.; Hacquard, T.; Derrien, B.; Izquierdo, E.; Lecorbeiller, M.; et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc. Natl. Acad. Sci. USA 2019, 116, 22872–22883. [Google Scholar] [CrossRef]
- Wu, J.; Michaeli, S.; Picchianti, L.; Dagdas, Y.; Galili, G.; Peled-Zehavi, H. ATI1 (ATG8-interacting protein 1) and ATI2 define a plant starvation-induced reticulophagy pathway and serve as MSBP1/MAPR5 cargo receptors. Autophagy 2021, 17, 3375–3388. [Google Scholar] [CrossRef]
- Fullgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014, 15, 65–74. [Google Scholar] [CrossRef]
- Fullgrabe, J.; Lynch-Day, M.A.; Heldring, N.; Li, W.; Struijk, R.B.; Ma, Q.; Hermanson, O.; Rosenfeld, M.G.; Klionsky, D.J.; Joseph, B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013, 500, 468–471. [Google Scholar] [CrossRef]
- He, M.; Xu, Y.; Chen, J.; Luo, Y.; Lv, Y.; Su, J.; Kershaw, M.J.; Li, W.; Wang, J.; Yin, J.; et al. MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 2018, 14, 1543–1561. [Google Scholar] [CrossRef]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J.; Chen, S.; Liu, S.; Li, Z.; Wang, Z.; Chen, B.; Zhang, C.; Zhang, Y.; Wu, J.; et al. Rhabdovirus encoded glycoprotein induces and harnesses host antiviral autophagy for maintaining its compatible infection. Autophagy 2024, 20, 275–294. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.Q. The hot issue: TOR signalling network in plants. Funct. Plant Biol. 2020, 48, 1–7. [Google Scholar] [CrossRef]
- Yuan, X.; Xu, P.; Yu, Y.; Xiong, Y. Glucose-TOR signaling regulates PIN2 stability to orchestrate auxin gradient and cell expansion in Arabidopsis root. Proc. Natl. Acad. Sci. USA 2020, 117, 32223–32225. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.Q. VvTOR interacts with VvSnRK1.1 and regulates sugar metabolism in grape. Planta 2022, 256, 56. [Google Scholar] [CrossRef]
- Caccamo, A.; Magri, A.; Medina, D.X.; Wisely, E.V.; Lopez-Aranda, M.F.; Silva, A.J.; Oddo, S. mTOR regulates tau phosphorylation and degradation: Implications for Alzheimer’s disease and other tauopathies. Aging Cell 2013, 12, 370–380. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Chang, Y.; Lim, G.; Huh, W.K. Analysis of the TORC1 interactome reveals a spatially distinct function of TORC1 in mRNP complexes. J. Cell Biol. 2021, 220, e201912060. [Google Scholar] [CrossRef]
- Bozulic, L.; Surucu, B.; Hynx, D.; Hemmings, B.A. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol. Cell 2008, 30, 203–213. [Google Scholar] [CrossRef]
- Bozulic, L.; Hemmings, B.A. PIKKing on PKB: Regulation of PKB activity by phosphorylation. Curr. Opin. Cell Biol. 2009, 21, 256–261. [Google Scholar] [CrossRef]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Zhang, C.; Wei, S.; Sun, W.P.; Teng, K.; Dai, M.M.; Wang, F.W.; Chen, J.W.; Ling, H.; Ma, X.D.; Feng, Z.H.; et al. Super-enhancer-driven AJUBA is activated by TCF4 and involved in epithelial-mesenchymal transition in the progression of Hepatocellular Carcinoma. Theranostics 2020, 10, 9066–9082. [Google Scholar] [CrossRef]
- Wang, Y.W.; Li, W.M.; Wu, W.J.; Chai, C.Y.; Liu, H.S.; Lai, M.D.; Chow, N.H. Potential significance of EMP3 in patients with upper urinary tract urothelial carcinoma: Crosstalk with ErbB2-PI3K-Akt pathway. J. Urol. 2014, 192, 242–251. [Google Scholar] [CrossRef]
- Hsieh, Y.H.; Hsieh, S.C.; Lee, C.H.; Yang, S.F.; Cheng, C.W.; Tang, M.J.; Lin, C.L.; Lin, C.L.; Chou, R.H. Targeting EMP3 suppresses proliferation and invasion of hepatocellular carcinoma cells through inactivation of PI3K/Akt pathway. Oncotarget 2015, 6, 34859–34874. [Google Scholar] [CrossRef]
- Christians, A.; Poisel, E.; Hartmann, C.; von Deimling, A.; Pusch, S. Characterization of the epithelial membrane protein 3 interaction network reveals a potential functional link to mitogenic signal transduction regulation. Int. J. Cancer 2019, 145, 461–473. [Google Scholar] [CrossRef]
- Sonoda, Y.; Warita, M.; Suzuki, T.; Ozawa, H.; Fukuda, Y.; Funakoshi-Tago, M.; Kasahara, T. Proteolipid protein 2 is associated with melanoma metastasis. Oncol. Rep. 2010, 23, 371–376. [Google Scholar] [CrossRef]
- Amaddii, M.; Meister, M.; Banning, A.; Tomasovic, A.; Mooz, J.; Rajalingam, K.; Tikkanen, R. Flotillin-1/reggie-2 protein plays dual role in activation of receptor-tyrosine kinase/mitogen-activated protein kinase signaling. J. Biol. Chem. 2012, 287, 7265–7278. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, F.; Liu, F.; Liu, X.; Huang, N.; Cai, X.; Sun, Y.; Li, A.; Luo, R. Overexpression of TIP30 inhibits the growth and invasion of glioma cells. Mol. Med. Rep. 2016, 13, 605–612. [Google Scholar] [CrossRef]
- Xu, G.; Dang, C. CMTM5 is downregulated and suppresses tumour growth in hepatocellular carcinoma through regulating PI3K-AKT signalling. Cancer Cell Int. 2017, 17, 113. [Google Scholar] [CrossRef]
- Wang, L.; Zhan, W.; Xie, S.; Hu, J.; Shi, Q.; Zhou, X.; Wu, Y.; Wang, S.; Fei, Z.; Yu, R. Over-expression of Rap2a inhibits glioma migration and invasion by down-regulating p-AKT. Cell Biol. Int. 2014, 38, 326–334. [Google Scholar] [CrossRef]
- Wu, J.X.; Du, W.Q.; Wang, X.C.; Wei, L.L.; Huo, F.C.; Pan, Y.J.; Wu, X.J.; Pei, D.S. Rap2a serves as a potential prognostic indicator of renal cell carcinoma and promotes its migration and invasion through up-regulating p-Akt. Sci. Rep. 2017, 7, 6623. [Google Scholar] [CrossRef]
- Arias, E.; Koga, H.; Diaz, A.; Mocholi, E.; Patel, B.; Cuervo, A.M. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol. Cell 2015, 59, 270–284. [Google Scholar] [CrossRef]
- Li, Y.; Xu, M.; Ding, X.; Yan, C.; Song, Z.; Chen, L.; Huang, X.; Wang, X.; Jian, Y.; Tang, G.; et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat. Cell Biol. 2016, 18, 1065–1077. [Google Scholar] [CrossRef]
- Matsuda-Lennikov, M.; Suizu, F.; Hirata, N.; Hashimoto, M.; Kimura, K.; Nagamine, T.; Fujioka, Y.; Ohba, Y.; Iwanaga, T.; Noguchi, M. Lysosomal interaction of Akt with Phafin2: A critical step in the induction of autophagy. PLoS ONE 2014, 9, e79795. [Google Scholar] [CrossRef]
- Hirata, N.; Suizu, F.; Matsuda-Lennikov, M.; Tanaka, T.; Edamura, T.; Ishigaki, S.; Donia, T.; Lithanatudom, P.; Obuse, C.; Iwanaga, T.; et al. Functional characterization of lysosomal interaction of Akt with VRK2. Oncogene 2018, 37, 5367–5386. [Google Scholar] [CrossRef]
- Amit, S.; Hatzubai, A.; Birman, Y.; Andersen, J.S.; Ben-Shushan, E.; Mann, M.; Ben-Neriah, Y.; Alkalay, I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002, 16, 1066–1076. [Google Scholar] [CrossRef]
- Bienz, M.; Clevers, H. Linking colorectal cancer to Wnt signaling. Cell 2000, 103, 311–320. [Google Scholar] [CrossRef]
- Polakis, P. Wnt signaling and cancer. Genes Dev. 2000, 14, 1837–1851. [Google Scholar] [CrossRef]
- Lee, E.; Salic, A.; Kruger, R.; Heinrich, R.; Kirschner, M.W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003, 1, E10. [Google Scholar] [CrossRef]
- Li, V.S.; Ng, S.S.; Boersema, P.J.; Low, T.Y.; Karthaus, W.R.; Gerlach, J.P.; Mohammed, S.; Heck, A.J.; Maurice, M.M.; Mahmoudi, T.; et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012, 149, 1245–1256. [Google Scholar] [CrossRef]
- Lybrand, D.B.; Naiman, M.; Laumann, J.M.; Boardman, M.; Petshow, S.; Hansen, K.; Scott, G.; Wehrli, M. Destruction complex dynamics: Wnt/β-catenin signaling alters Axin-GSK3β interactions in vivo. Development 2019, 146, dev164145. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4, a007906. [Google Scholar] [CrossRef]
- Yu, R.; Guo, X.; Zhong, J.; Li, M.; Zeng, Z.; Zhang, H. The N-terminal HSDCIF motif is required for cell surface trafficking and dimerization of family B G protein coupled receptor PAC1. PLoS ONE 2012, 7, e51811. [Google Scholar] [CrossRef]
- Yu, R.; Cui, Z.; Li, M.; Yang, Y.; Zhong, J. Dimer-dependent intrinsic/basal activity of the class B G protein-coupled receptor PAC1 promotes cellular anti-apoptotic activity through Wnt/β-catenin pathways that are associated with dimer endocytosis. PLoS ONE 2014, 9, e113913. [Google Scholar] [CrossRef]
- Carron, C.; Pascal, A.; Djiane, A.; Boucaut, J.C.; Shi, D.L.; Umbhauer, M. Frizzled receptor dimerization is sufficient to activate the Wnt/beta-catenin pathway. J. Cell Sci. 2003, 116 Pt 12, 2541–2550. [Google Scholar] [CrossRef]
- Green, J.L.; Inoue, T.; Sternberg, P.W. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development 2007, 134, 4053–4062. [Google Scholar] [CrossRef]
- Hayashi, Y.; Hirotsu, T.; Iwata, R.; Kage-Nakadai, E.; Kunitomo, H.; Ishihara, T.; Iino, Y.; Kubo, T. A trophic role for Wnt-Ror kinase signaling during developmental pruning in Caenorhabditis elegans. Nat. Neurosci. 2009, 12, 981–987. [Google Scholar] [CrossRef]
- Modzelewska, K.; Lauritzen, A.; Hasenoeder, S.; Brown, L.; Georgiou, J.; Moghal, N. Neurons refine the Caenorhabditis elegans body plan by directing axial patterning by Wnts. PLoS Biol. 2013, 11, e1001465. [Google Scholar] [CrossRef]
- Pandey, P.; Bhardwaj, A.; Babu, K. Regulation of WNT Signaling at the Neuromuscular Junction by the Immunoglobulin Superfamily Protein RIG-3 in Caenorhabditis elegans. Genetics 2017, 206, 1521–1534. [Google Scholar] [CrossRef]
- Moynagh, P.N. The NF-kappaB pathway. J. Cell Sci. 2005, 118 Pt 20, 4589–4592. [Google Scholar] [CrossRef]
- Karin, M.; Lin, A. NF-kappaB at the crossroads of life and death. Nat. Immunol. 2002, 3, 221–227. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef]
- Dorsey, M.J.; Tae, H.J.; Sollenberger, K.G.; Mascarenhas, N.T.; Johansen, L.M.; Taparowsky, E.J. B-ATF: A novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 1995, 11, 2255–2265. [Google Scholar]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, R.; Fu, P.; Chen, L.; Luo, L.; Chu, P.; He, L.; Li, Y.; Liao, L.; Zhu, Z.; et al. Investigating the Role of BATF3 in Grass Carp (Ctenopharyngodon idella) Immune Modulation: A Fundamental Functional Analysis. Int. J. Mol. Sci. 2019, 20, 1687. [Google Scholar] [CrossRef]
- Zhou, X.Y.; Hu, D.X.; Chen, R.Q.; Chen, X.Q.; Dong, W.L.; Yi, C.L. 14-3-3 Isoforms Differentially Regulate NFκB Signaling in the Brain After Ischemia-Reperfusion. Neurochem. Res. 2017, 42, 2354–2362. [Google Scholar] [CrossRef]
- Miyakura, H.; Fukuda, M.; Enomoto, H.; Ishikawa, K.; Watanabe, S.; Semba, K. A screening system for identifying interacting proteins using biomolecular fluorescence complementation and transposon gene trap. PLoS ONE 2021, 16, e0251240. [Google Scholar] [CrossRef]
- Talaty, P.; Emery, A.; Holthusen, K.; Everly, D.N., Jr. Identification of transmembrane protein 134 as a novel LMP1-binding protein by using bimolecular fluorescence complementation and an enhanced retroviral mutagen. J. Virol. 2012, 86, 11345–11355. [Google Scholar] [CrossRef]
- Chen, H.; Cai, X.; Li, R.; Wu, Y.; Qiu, H.; Zheng, J.; Zhou, D.; Fang, J.; Wu, X. A novel toll-like receptor from Crassostrea gigas is involved in innate immune response to Vibrio alginolyticus. Infect. Genet. Evol. 2022, 97, 105159. [Google Scholar] [CrossRef]
- Group, M. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Pedley, K.F.; Martin, G.B. Role of mitogen-activated protein kinases in plant immunity. Curr. Opin. Plant Biol. 2005, 8, 541–547. [Google Scholar] [CrossRef]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef]
- Suarez-Rodriguez, M.C.; Adams-Phillips, L.; Liu, Y.; Wang, H.; Su, S.H.; Jester, P.J.; Zhang, S.; Bent, A.F.; Krysan, P.J. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007, 143, 661–669. [Google Scholar] [CrossRef]
- Gao, M.; Liu, J.; Bi, D.; Zhang, Z.; Cheng, F.; Chen, S.; Zhang, Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008, 18, 1190–1198. [Google Scholar] [CrossRef]
- Owens, D.M.; Keyse, S.M. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007, 26, 3203–3213. [Google Scholar] [CrossRef]
- Lumbreras, V.; Vilela, B.; Irar, S.; Sole, M.; Capellades, M.; Valls, M.; Coca, M.; Pages, M. MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 2010, 63, 1017–1030. [Google Scholar] [CrossRef]
- Wang, H.; Gong, W.; Wang, Y.; Ma, Q. Contribution of a WRKY Transcription Factor, ShWRKY81, to Powdery Mildew Resistance in Wild Tomato. Int. J. Mol. Sci. 2023, 24, 2583. [Google Scholar] [CrossRef]
- Wang, F.; Liang, S.; Wang, G.; Wang, Q.; Xu, Z.; Li, B.; Fu, C.; Fan, Y.; Hu, T.; Alariqi, M.; et al. Comprehensive analysis of MAPK gene family in upland cotton (Gossypium hirsutum) and functional characterization of GhMPK31 in regulating defense response to insect infestation. Plant Cell Rep. 2024, 43, 102. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef]
- Yamada, K.; Yamaguchi, K.; Yoshimura, S.; Terauchi, A.; Kawasaki, T. Conservation of Chitin-Induced MAPK Signaling Pathways in Rice and Arabidopsis. Plant Cell Physiol. 2017, 58, 993–1002. [Google Scholar] [CrossRef]
- de Zelicourt, A.; Colcombet, J.; Hirt, H. The Role of MAPK Modules and ABA during Abiotic Stress Signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Jonak, C.; Okresz, L.; Bogre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, C.; Yang, B.; Wu, F.; Hao, X.; Liang, W.; Niu, F.; Yan, J.; Zhang, H.; Wang, B.; et al. Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J. Exp. Bot. 2014, 65, 2171–2188. [Google Scholar] [CrossRef]
- Chen, L.; Sun, H.; Wang, F.; Yue, D.; Shen, X.; Sun, W.; Zhang, X.; Yang, X. Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14-GhMKK11-GhMPK31 pathway is involved in the drought response in cotton. Plant Mol. Biol. 2020, 103, 211–223. [Google Scholar] [CrossRef]
- Jung, W.J.; Yoon, J.S.; Seo, Y.W. TaMAPK3 phosphorylates TaCBF and TaICE and plays a negative role in wheat freezing tolerance. J. Plant Physiol. 2024, 296, 154233. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Sharma, M.; Negi, S.; Kumar, P.; Srivastava, D.K.; Choudhary, M.K.; Irfan, M. Fruit ripening under heat stress: The intriguing role of ethylene-mediated signaling. Plant Sci. 2023, 335, 111820. [Google Scholar] [CrossRef]
- Song, Z.; Qin, J.; Zheng, Q.; Ding, X.; Chen, W.; Lu, W.; Li, X.; Zhu, X. The Involvement of the Banana F-Box Protein MaEBF1 in Regulating Chilling-Inhibited Starch Degradation through Interaction with a MaNAC67-Like Protein. Biomolecules 2019, 9, 552. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Qiao, L.; Chen, J.; Pappa, M.B.; Pei, H.; Zhang, T.; Chang, C.; Dong, C.H. Arabidopsis CPR5 regulates ethylene signaling via molecular association with the ETR1 receptor. J. Integr. Plant Biol. 2017, 59, 810–824. [Google Scholar] [CrossRef]
- Chen, J.; Sui, X.; Ma, B.; Li, Y.; Li, N.; Qiao, L.; Yu, Y.; Dong, C.H. Arabidopsis CPR5 plays a role in regulating nucleocytoplasmic transport of mRNAs in ethylene signaling pathway. Plant Cell Rep. 2022, 41, 1075–1085. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Yang, Y.; Li, Z.; Li, H.; Yu, R.; Luan, F.; Zhang, X.; Wei, C. Novel Bisexual Flower Control Gene Regulates Sex Differentiation in Melon (Cucumis melo L.). J. Agric. Food Chem. 2022, 70, 15401–15414. [Google Scholar] [CrossRef]
- Abiri, R.; Shaharuddin, N.A.; Maziah, M.; Yusof, Z.N.B.; Atabaki, N.; Sahebi, M.; Valdiani, A.; Kalhori, N.; Azizi, P.; Hanafi, M.M. Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 2017, 134, 33–44. [Google Scholar] [CrossRef]
- Cheng, H.; Yu, Y.; Zhai, Y.; Wang, L.; Wang, L.; Chen, S.; Chen, F.; Jiang, J. An ethylene-responsive transcription factor and a B-box protein coordinate vegetative growth and photoperiodic flowering in chrysanthemum. Plant Cell Environ. 2023, 46, 440–450. [Google Scholar] [CrossRef]
- Du, X.; Fang, L.; Li, J.; Chen, T.; Cheng, Z.; Zhu, B.; Gu, L.; Wang, H. The TabHLH094-TaMYC8 complex mediates the cadmium response in wheat. Mol. Breed. 2023, 43, 57. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, S.; Yang, G.; Li, L.; Li, J.; Yang, F. The MYB transcription factor SmMYB113 directly regulates ethylene-dependent flower abscission in eggplant. Plant Physiol. Biochem. 2024, 209, 108544. [Google Scholar] [CrossRef]
- Cheng, M.C.; Liao, P.M.; Kuo, W.W.; Lin, T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef]
- Poór, P.; Nawaz, K.; Gupta, R.; Ashfaque, F.; Khan, M.I.R. Ethylene involvement in the regulation of heat stress tolerance in plants. Plant Cell Rep. 2022, 41, 675–698. [Google Scholar] [CrossRef]
- Li, N.; Euring, D.; Cha, J.Y.; Lin, Z.; Lu, M.; Huang, L.J.; Kim, W.Y. Plant Hormone-Mediated Regulation of Heat Tolerance in Response to Global Climate Change. Front. Plant Sci. 2020, 11, 627969. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Wang, R.; Xu, F.; Tong, S.; Song, C.; Shao, Y.; Yi, M.; He, J. Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum). Int. J. Mol. Sci. 2022, 23, 16135. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.; Zhou, Y.; Chen, J.; Xu, Z.; Ma, R.; Dong, Y.; Ma, Y.; Chen, M. AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybean by interacting with GmERFs. Plant Physiol. Biochem. 2022, 170, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Voβ, U.; Farcot, E.; Bennett, M.J.; Bishopp, A. Systems biology approaches to understand the role of auxin in root growth and development. Physiol. Plant 2014, 151, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Wang, X.J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.; Yu, J.; Huang, S.; Zhang, Y.; Wei, H.; Wei, Z. Genome-Wide Identification and Characterization of Auxin Response Factor (ARF) Gene Family Involved in Wood Formation and Response to Exogenous Hormone Treatment in Populus trichocarpa. Int. J. Mol. Sci. 2023, 24, 740. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Chong, X.; Guan, Y.; Jiang, J.; Zhang, F.; Wang, H.; Song, A.; Chen, S.; Ding, L.; Chen, F. Heterologous expression of chrysanthemum TOPLESS corepressor CmTPL1-1 alters meristem maintenance and organ development in Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 157, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Ke, Y.; Zhou, Y.; Yu, Y.; Waseem, M.; Ashraf, U.; Li, X.; Yu, R.; Fan, Y. Genome-wide analysis of ARF transcription factors reveals HcARF5 expression profile associated with the biosynthesis of beta-ocimene synthase in Hedychium coronarium. Plant Cell Rep. 2021, 40, 1269–1284. [Google Scholar] [CrossRef]
- Gonzalez-Lamothe, R.; El Oirdi, M.; Brisson, N.; Bouarab, K. The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 2012, 24, 762–777. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; Zhai, Y.; Kamal, H.; Smertenko, A.; Pappu, H.R. A plant virus protein, NIa-pro, interacts with Indole-3-acetic acid-amido synthetase, whose levels positively correlate with disease severity. Front. Plant Sci. 2023, 14, 1112821. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Xu, Z.; Kong, Y. Overexpression of NtEXPA11 modulates plant growth and development and enhances stress tolerance in tobacco. Plant Physiol. Biochem. 2020, 151, 477–485. [Google Scholar] [CrossRef]
- Sun, W.; Yao, M.; Wang, Z.; Chen, Y.; Zhan, J.; Yan, J.; Jiang, S.; Jian, S.; Chen, H.; Bu, T.; et al. Involvement of Auxin-Mediated CqEXPA50 Contributes to Salt Tolerance in Quinoa (Chenopodium quinoa) by Interaction with Auxin Pathway Genes. Int. J. Mol. Sci. 2022, 23, 8480. [Google Scholar] [CrossRef]
- Song, Y.; Li, S.; Sui, Y.; Zheng, H.; Han, G.; Sun, X.; Yang, W.; Wang, H.; Zhuang, K.; Kong, F.; et al. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum. Theor. Appl. Genet. 2022, 135, 201–216. [Google Scholar] [CrossRef]
- Garmendia, A.; Beltran, R.; Zornoza, C.; Garcia-Breijo, F.J.; Reig, J.; Merle, H. Gibberellic acid in Citrus spp. flowering and fruiting: A systematic review. PLoS ONE 2019, 14, e0223147. [Google Scholar] [CrossRef]
- Camara, M.C.; Vandenberghe, L.P.S.; Rodrigues, C.; de Oliveira, J.; Faulds, C.; Bertrand, E.; Soccol, C.R. Current advances in gibberellic acid (GA3) production, patented technologies and potential applications. Planta 2018, 248, 1049–1062. [Google Scholar] [CrossRef]
- van der Schoot, C.; Paul, L.K.; Rinne, P.L. The embryonic shoot: A lifeline through winter. J. Exp. Bot. 2014, 65, 1699–1712. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Zhang, Z.; Liu, W.; Qiu, L.; Wang, J.; Cheng, T.; Zhang, Q. Integrative Identification of Crucial Genes Associated With Plant Hormone-Mediated Bud Dormancy in Prunus mume. Front. Genet. 2021, 12, 698598. [Google Scholar] [CrossRef]
- Yuxi, Z.; Yanchao, Y.; Zejun, L.; Tao, Z.; Feng, L.; Chunying, L.; Shupeng, G. GA3 is superior to GA4 in promoting bud endodormancy release in tree peony (Paeonia suffruticosa) and their potential working mechanism. BMC Plant Biol. 2021, 21, 323. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, G.L.; Song, S.Y.; Zhao, Y.N.; Lu, S.; Zhou, F. ORANGE negatively regulates flowering time in Arabidopsisthaliana. J. Plant Physiol. 2022, 274, 153719. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, L. Epigenetic Regulation of Gibberellin Metabolism and Signaling. Plant Cell Physiol. 2020, 61, 1912–1918. [Google Scholar] [CrossRef]
- Acheampong, A.K.; Hu, J.; Rotman, A.; Zheng, C.; Halaly, T.; Takebayashi, Y.; Jikumaru, Y.; Kamiya, Y.; Lichter, A.; Sun, T.P.; et al. Functional characterization and developmental expression profiling of gibberellin signalling components in Vitis vinifera. J. Exp. Bot. 2015, 66, 1463–1476. [Google Scholar] [CrossRef]
- Li, S.; Wang, Q.; Wen, B.; Zhang, R.; Jing, X.; Xiao, W.; Chen, X.; Tan, Q.; Li, L. Endodormancy Release Can Be Modulated by the GA4-GID1c-DELLA2 Module in Peach Leaf Buds. Front. Plant Sci. 2021, 12, 713514. [Google Scholar] [CrossRef]
- Lv, L.; Huo, X.; Wen, L.; Gao, Z.; Khalil-Ur-Rehman, M. Isolation and Role of PmRGL2 in GA-mediated Floral Bud Dormancy Release in Japanese Apricot (Prunus mume Siebold et Zucc.). Front. Plant Sci. 2018, 9, 27. [Google Scholar] [CrossRef]
- Chai, Z.; Fang, J.; Huang, C.; Huang, R.; Tan, X.; Chen, B.; Yao, W.; Zhang, M. A novel transcription factor, ScAIL1, modulates plant defense responses by targeting DELLA and regulating gibberellin and jasmonic acid signaling in sugarcane. J. Exp. Bot. 2022, 73, 6727–6743. [Google Scholar] [CrossRef]
- Gao, L.; Niu, D.; Chi, T.; Yuan, Y.; Liu, C.; Gai, S.; Zhang, Y. PsRGL1 negatively regulates chilling- and gibberellin-induced dormancy release by PsF-box1-mediated targeting for proteolytic degradation in tree peony. Hortic. Res. 2023, 10, uhad044. [Google Scholar] [CrossRef]
- Ou, X.; Wang, Y.; Li, J.; Zhang, J.; Xie, Z.; He, B.; Jiang, Z.; Wang, Y.; Su, W.; Song, S.; et al. Genome-wide identification of the KNOTTED HOMEOBOX gene family and their involvement in stalk development in flowering Chinese cabbage. Front. Plant Sci. 2022, 13, 1019884. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J.; Xia, L.; Chen, C.; Wang, M.; Lu, J.; Lu, T.; Li, K.; Liang, R.; He, X.; et al. Functional studies of four MiFPF genes in mango revealed their function in promoting flowering in transgenic Arabidopsis. J. Plant Physiol. 2023, 285, 153994. [Google Scholar] [CrossRef]
- Hu, J.; Israeli, A.; Ori, N.; Sun, T.P. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef]
- Li, J.; Gong, J.; Zhang, L.; Shen, H.; Chen, G.; Xie, Q.; Hu, Z. Overexpression of SlPRE5, an atypical bHLH transcription factor, affects plant morphology and chlorophyll accumulation in tomato. J. Plant Physiol. 2022, 273, 153698. [Google Scholar] [CrossRef]
- Guo, P.; Yang, Q.; Wang, Y.; Yang, Z.; Xie, Q.; Chen, G.; Chen, X.; Hu, Z. Overexpression of SlPRE3 alters the plant morphologies in Solanum lycopersicum. Plant Cell Rep. 2023, 42, 1907–1925. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Yan, Y.; Zhang, S.; Li, H.; Gao, Z.; Wang, C.; Guo, X. GhWRKY21 regulates ABA-mediated drought tolerance by fine-tuning the expression of GhHAB in cotton. Plant Cell Rep. 2021, 40, 2135–2150. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Chu, Y.; Wang, Y.; Diao, Y.; Zhao, Y.; Liu, L.; Wei, X.; Meng, Y.; Li, F.; Ge, X. The miR164-GhCUC2-GhBRC1 module regulates plant architecture through abscisic acid in cotton. Plant Biotechnol. J. 2021, 19, 1839–1851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Ye, T.; Lu, Y.; Chen, X.; Wu, Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013, 64, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Li, G.; Wang, H.; Zhao, J.; Li, Z.; Liu, X.; Shu, Y.; Liu, W.; Wang, S.; Huang, J.; et al. Exogenous abscisic acid represses rice flowering via SAPK8-ABF1-Ehd1/Ehd2 pathway. J. Adv. Res. 2024, 59, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Zhao, Z.Q.; Song, D.D.; Yuan, Y.X.; Sun, H.J.; Zhao, J.F.; Chen, Y.L.; Zhang, C.G. SnRK2.6 interacts with phytochrome B and plays a negative role in red light-induced stomatal opening. Plant Signal. Behav. 2021, 16, 1913307. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, L. ALA Upregulates MdPTPA Expression to Increase the PP2A Activity and Promote Stomatal Opening in Apple Leaves. Plant Sci. 2022, 325, 111490. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; An, Y.; Wang, L. ALA reverses ABA-induced stomatal closure by modulating PP2AC and SnRK2.6 activity in apple leaves. Hortic. Res. 2023, 10, uhad067. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, M.; Wang, L.; Sun, W.; Li, M.; Feng, C.; Yang, X. Genome-wide identification and expression analysis of PYL family genes and functional characterization of GhPYL8D2 under drought stress in Gossypium hirsutum. Plant Physiol. Biochem. 2023, 203, 108072. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Hou, X.; Ni, C.; Han, L.; Du, P.; Xiao, K. Wheat ABA Receptor TaPYL5 Constitutes a Signaling Module with Its Downstream Partners TaPP2C53/TaSnRK2.1/TaABI1 to Modulate Plant Drought Response. Int. J. Mol. Sci. 2023, 24, 7969. [Google Scholar] [CrossRef]
- Liu, S.; Lu, C.; Jiang, G.; Zhou, R.; Chang, Y.; Wang, S.; Wang, D.; Niu, J.; Wang, Z. Comprehensive functional analysis of the PYL-PP2C-SnRK2s family in Bletilla striata reveals that BsPP2C22 and BsPP2C38 interact with BsPYLs and BsSnRK2s in response to multiple abiotic stresses. Front. Plant Sci. 2022, 13, 963069. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Jiang, Z.; Wei, X.; Liu, S.; Qu, J.; Guan, S.; Ma, Y. Overexpression of the homeobox-leucine zipper protein ATHB-6 improves the drought tolerance of maize (Zea mays L.). Plant Sci. Int. J. Exp. Plant Biol. 2022, 316, 111159. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Jiang, Z.; Miao, M.; Wei, X.; Wang, C.; Liu, S.; Guan, S.; Ma, Y. Zmhdz9, an HD-Zip transcription factor, promotes drought stress resistance in maize by modulating ABA and lignin accumulation. Int. J. Biol. Macromol. 2023, 258 Pt 1, 128849. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.D.; Koraichi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S. A new protein-protein interaction sensor based on tripartite split-GFP association. Sci. Rep. 2013, 3, 2854. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lin, J.; Zhou, C.; Deng, X.; Xia, B. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta Biochim. Biophys. Sin. 2011, 43, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Ying, Y.; Ma, Z.; Yang, Y.; Chen, A.K. A Background Assessable and Correctable Bimolecular Fluorescence Complementation System for Nanoscopic Single-Molecule Imaging of Intracellular Protein-Protein Interactions. ACS Nano 2021, 15, 14338–14346. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, A.; Huang, T.; Lin, L.J.; Nan, X. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS ONE 2014, 9, e100589. [Google Scholar] [CrossRef] [PubMed]
- Hertel, F.; Mo, G.C.; Duwe, S.; Dedecker, P.; Zhang, J. RefSOFI for Mapping Nanoscale Organization of Protein-Protein Interactions in Living Cells. Cell Rep. 2016, 14, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xing, D.; Su, Q.P.; Zhu, Y.; Zhang, J.; Kong, X.; Xue, B.; Wang, S.; Sun, H.; Tao, Y.; et al. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nat. Commun. 2014, 5, 4443. [Google Scholar] [CrossRef]
- Wang, S.; Ding, M.; Chen, X.; Chang, L.; Sun, Y. Development of bimolecular fluorescence complementation using rsEGFP2 for detection and super-resolution imaging of protein-protein interactions in live cells. Biomed. Opt. Express 2017, 8, 3119–3131. [Google Scholar] [CrossRef]
- Chen, M.; Liu, S.; Li, W.; Zhang, Z.; Zhang, X.; Zhang, X.E.; Cui, Z. Three-Fragment Fluorescence Complementation Coupled with Photoactivated Localization Microscopy for Nanoscale Imaging of Ternary Complexes. ACS Nano 2016, 10, 8482–8490. [Google Scholar] [CrossRef]
- Lu, K.; Vu, C.Q.; Matsuda, T.; Nagai, T. Fluorescent Protein-Based Indicators for Functional Super-Resolution Imaging of Biomolecular Activities in Living Cells. Int. J. Mol. Sci. 2019, 20, 5784. [Google Scholar] [CrossRef]
- Jiang, Y.; Peng, J.; Cao, Y.; Han, Z.; Zhang, L.; Su, W.; Lin, S.; Yuan, Y.; Wang, B.; Yang, X.; et al. Method for fast staining and obtaining high-magnification and high-resolution cell images of Nicotiana benthamiana. Physiol. Mol. Biol. Plants 2021, 27, 181–188. [Google Scholar] [CrossRef]
- Zilian, E.; Maiss, E. An optimized mRFP-based bimolecular fluorescence complementation system for the detection of protein-protein interactions in planta. J. Virol. Methods 2011, 174, 158–165. [Google Scholar] [CrossRef]
- Castillo, S.; Gence, R.; Pagan, D.; Koraïchi, F.; Bouchenot, C.; Pons, B.J.; Boëlle, B.; Olichon, A.; Lajoie-Mazenc, I.; Favre, G.; et al. Visualizing the subcellular localization of RHOB-GTP and GTPase-Effector complexes using a split-GFP/nanobody labelling assay. Eur. J. Cell Biol. 2023, 102, 151355. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, W.; Gan, C.; Wei, X.; Li, J.; Sun, Y.; Yuan, Y.; Cai, X.; Long, Q.; Cui, J.; et al. Strategies to improve the fluorescent signal of the tripartite sfGFP system. Acta Biochim. Biophys. Sin. 2020, 52, 998–1006. [Google Scholar] [CrossRef]
- Mazza, D.; Abernathy, A.; Golob, N.; Morisaki, T.; McNally, J.G. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012, 40, e119. [Google Scholar] [CrossRef]
- Shao, S.; Xue, B.; Sun, Y. Intranucleus Single-Molecule Imaging in Living Cells. Biophys. J. 2018, 115, 181–189. [Google Scholar] [CrossRef]
- Shao, S.; Zhang, H.; Zeng, Y.; Li, Y.; Sun, C.; Sun, Y. TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells. Commun. Biol. 2021, 4, 378. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Suk, H.J.; Kodandaramaiah, S.B.; Yoshida, F.; DeGennaro, E.M.; Drobizhev, M.; Hughes, T.E.; Desimone, R.; Boyden, E.S.; Verkhusha, V.V. Near-Infrared Fluorescent Proteins Engineered from Bacterial Phytochromes in Neuroimaging. Biophys. J. 2017, 113, 2299–2309. [Google Scholar] [CrossRef]
- Liu, F.; Hu, H.; Deng, M.; Xiang, Z.; Guo, Y.; Guan, X.; Li, D.; Hu, Q.; Lei, W.; Peng, H.; et al. A Bright Monomeric Near-Infrared Fluorescent Protein with an Excitation Peak at 633 nm for Labeling Cellular Protein and Reporting Protein-Protein Interaction. ACS Sens. 2022, 7, 1855–1866. [Google Scholar] [CrossRef] [PubMed]
- Valli, J.; Garcia-Burgos, A.; Rooney, L.M.; Vale de Melo, E.O.B.; Duncan, R.R.; Rickman, C. Seeing beyond the limit: A guide to choosing the right super-resolution microscopy technique. J. Biol. Chem. 2021, 297, 100791. [Google Scholar] [CrossRef]
- Ortiz-Vilchis, P.; De-la-Cruz-Garcia, J.S.; Ramirez-Arellano, A. Identification of Relevant Protein Interactions with Partial Knowledge: A Complex Network and Deep Learning Approach. Biology 2023, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Wilson, E.L.; Yu, Y.; Leal, N.S.; Woodward, J.A.; Patikas, N.; Morris, J.L.; Field, S.F.; Plumbly, W.; Paupe, V.; Chowdhury, S.R.; et al. Genome-wide CRISPR/Cas9 screen shows that loss of GET4 increases mitochondria-endoplasmic reticulum contact sites and is neuroprotective. Cell Death Dis. 2024, 15, 203. [Google Scholar] [CrossRef]
- Hong, Y.; Lu, G.; Duan, J.; Liu, W.; Zhang, Y. Comparison and optimization of CRISPR/dCas9/gRNA genome-labeling systems for live cell imaging. Genome Biol. 2018, 19, 39. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).