Characterization of Subcellular Dynamics of Sterol Methyltransferases Clarifies Defective Cell Division in smt2 smt3, a C-24 Ethyl Sterol-Deficient Mutant of Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Expression Plasmids
2.3. Plant Transformation
2.4. Transformation of Tobacco BY-2 Cells
2.5. Imaging Analyses
2.6. Sterol Analysis
3. Results
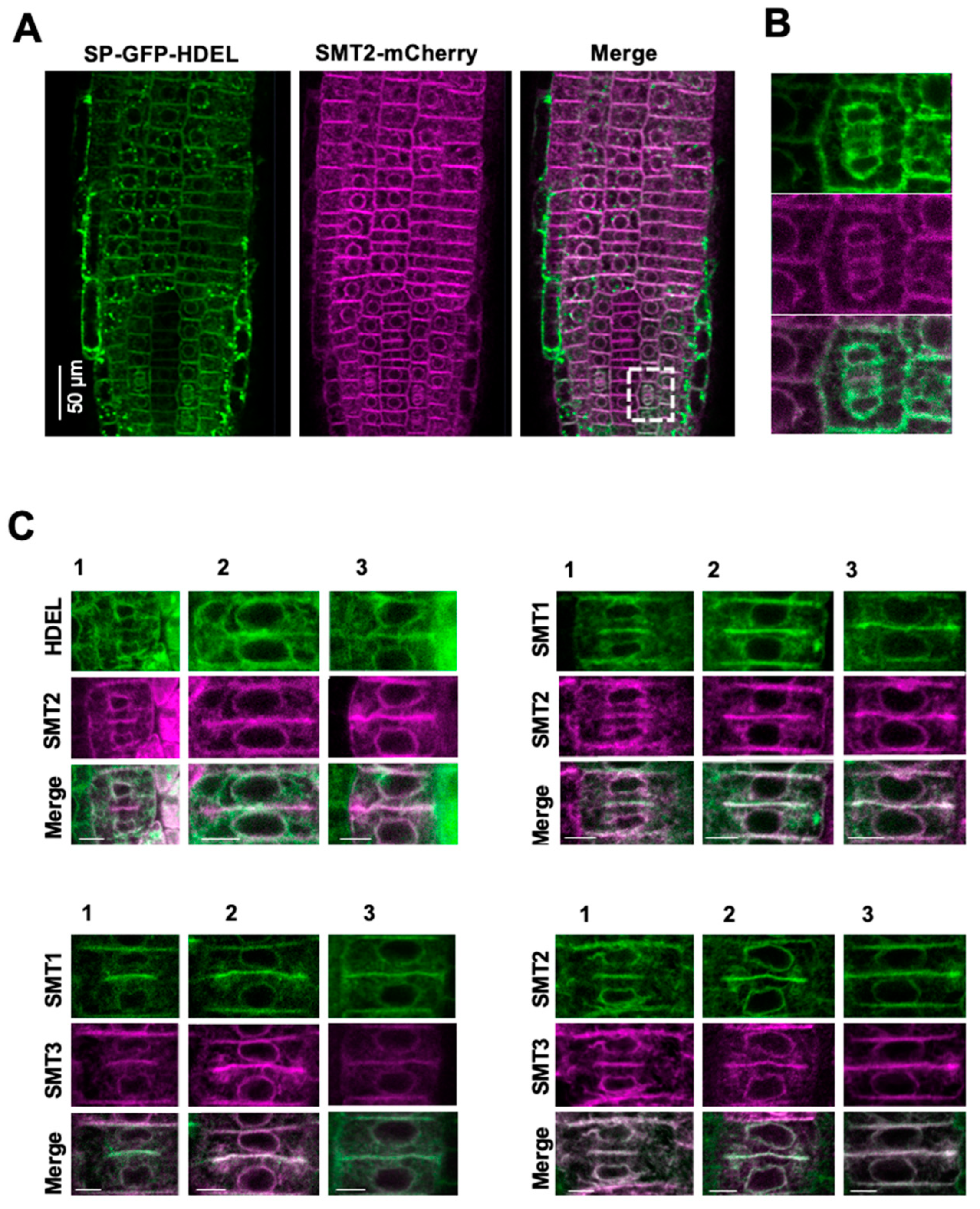
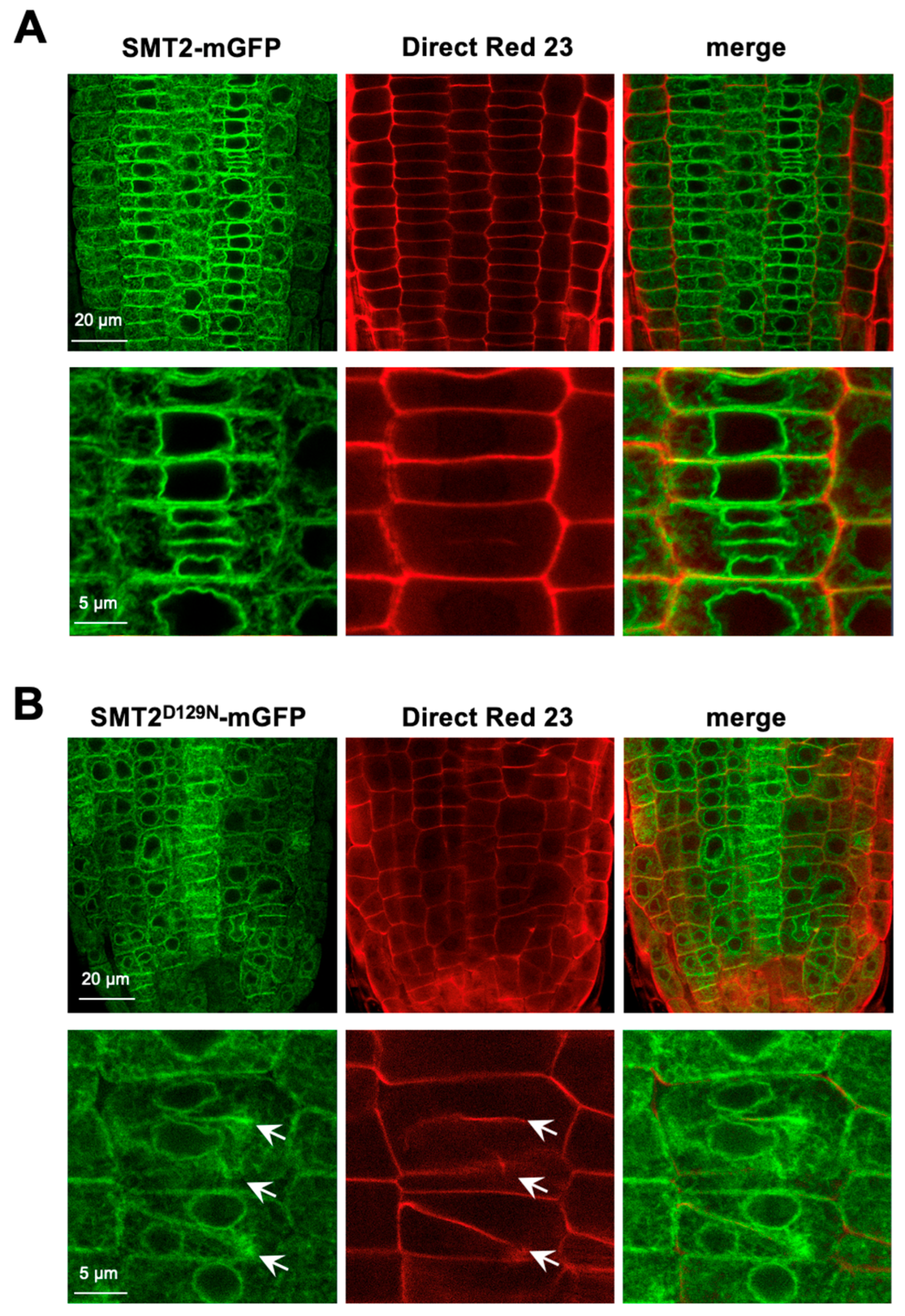
3.1. Subcellular Localizations of Sterol Biosynthetic Enzymes
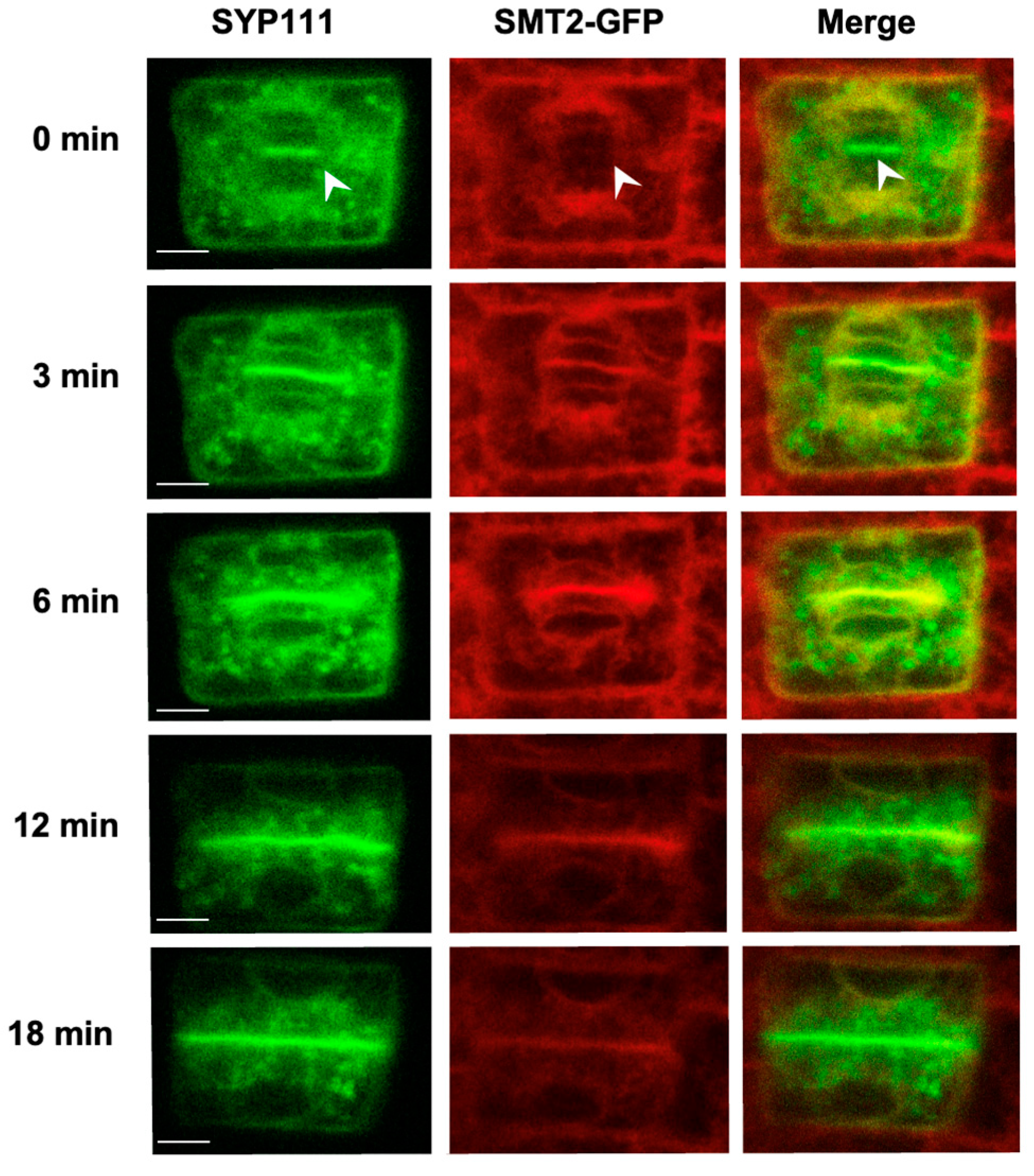
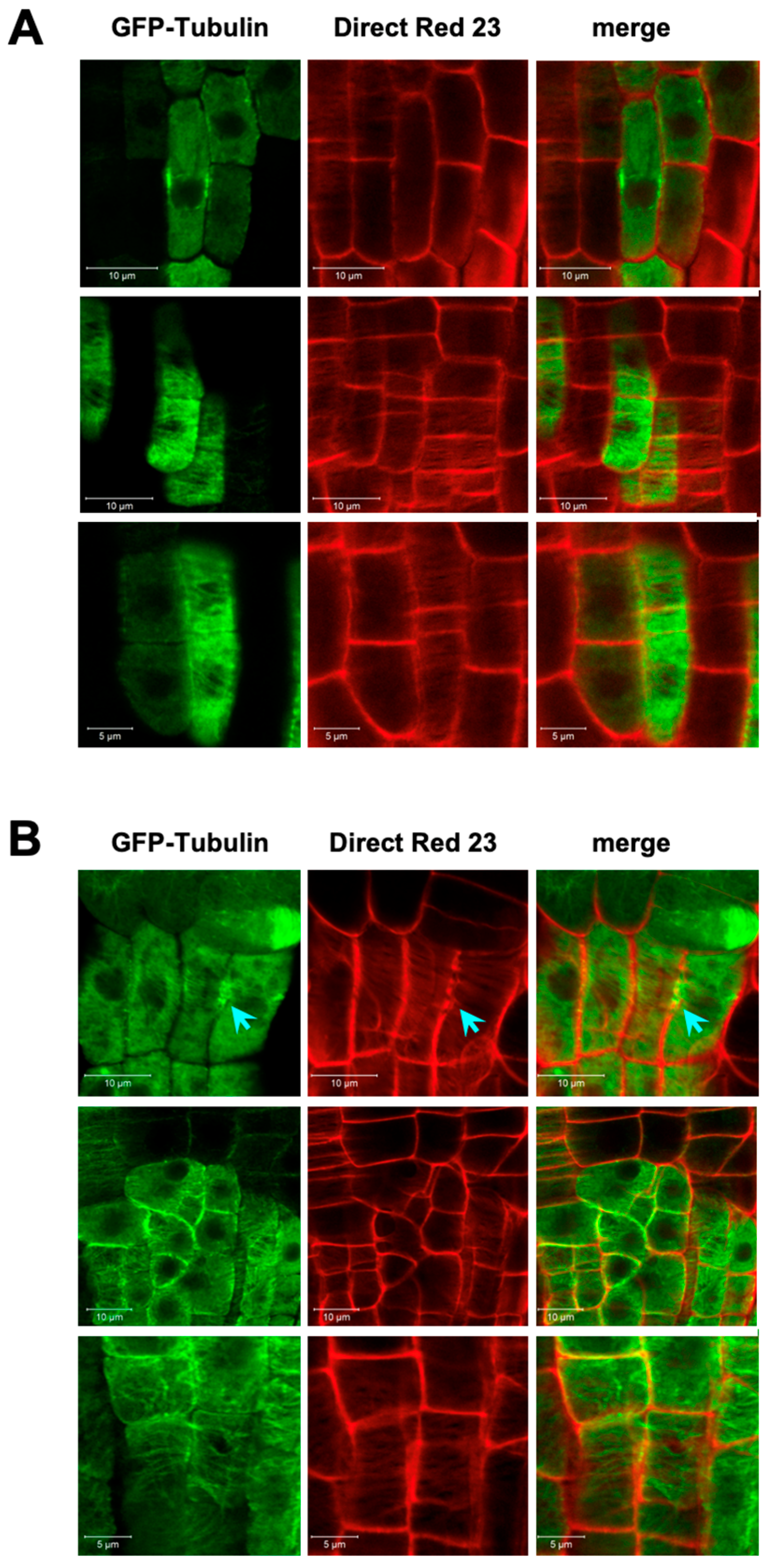
3.2. Endomembrane System and Cell Plate Assembly
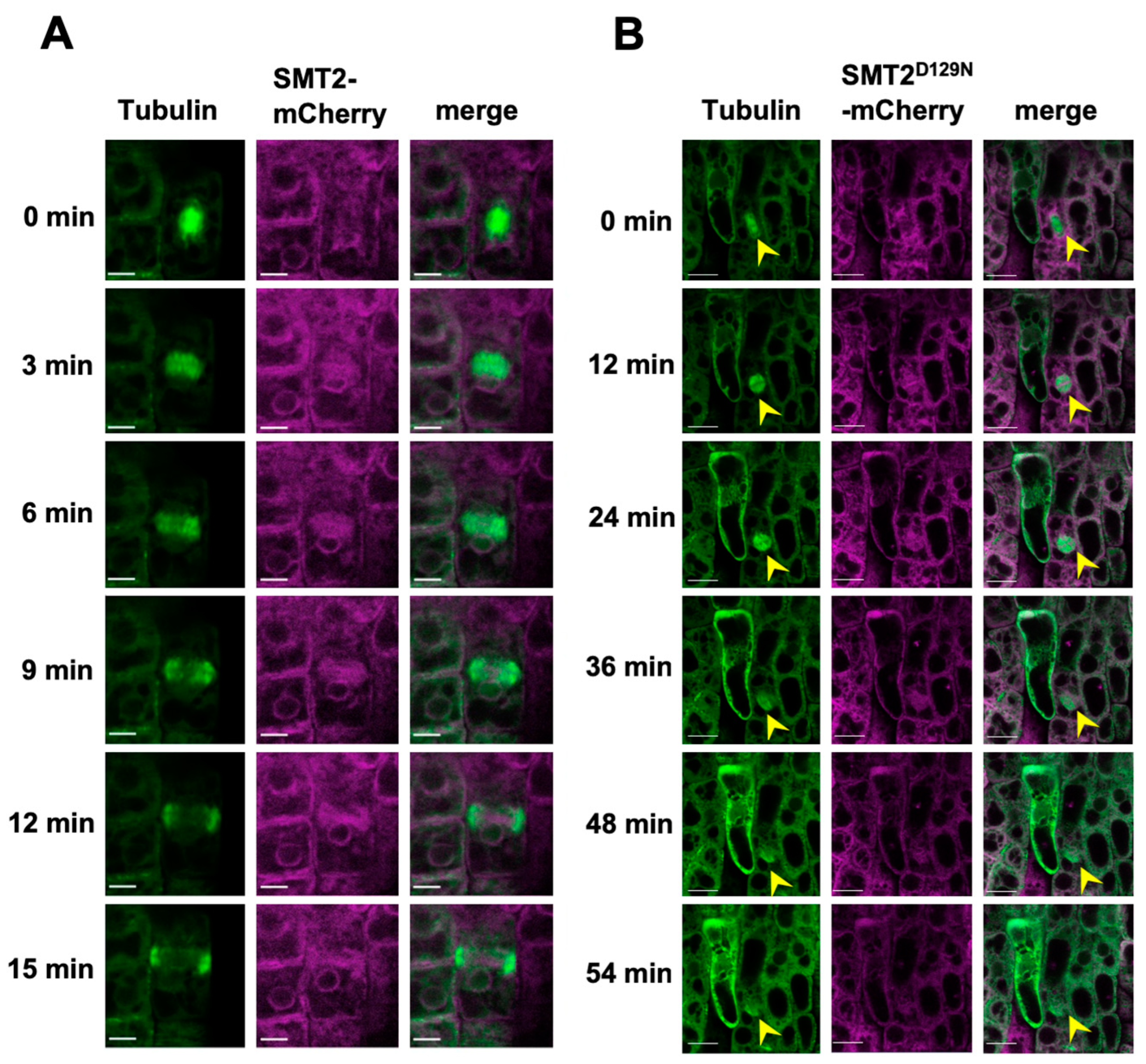
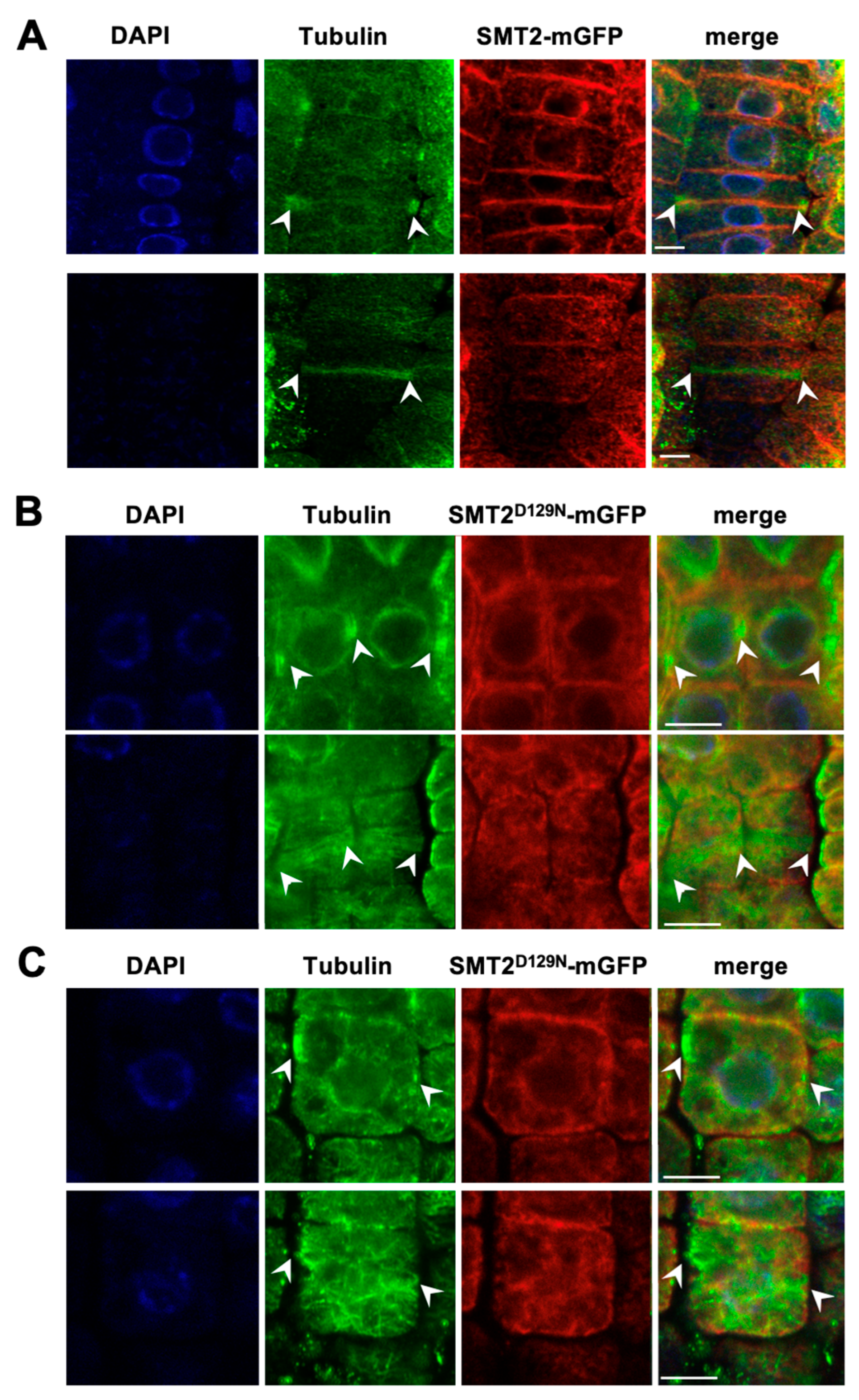
3.3. Cell Division Plane
4. Discussion
4.1. Subcellular Sites of Sterol Biosynthesis
4.2. Defected Cell Division
4.3. Critical Roles of C-24 Ethyl Sterols
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schaller, H. Sterol and steroid biosynthesis and metabolism in plants and microorganisms. In Comprehensive Natural Products II; Hung-Wen, L., Mander, L., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2010; pp. 755–787. [Google Scholar]
- Nes, W.D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451. [Google Scholar] [CrossRef] [PubMed]
- Moreau, R.A.; Nystrom, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef]
- Darnet, S.; Schaller, H. Metabolism and biological activities of 4-methyl-sterols. Molecules 2019, 24, 451. [Google Scholar] [CrossRef]
- Hartmann, M.-A.; Benveniste, P. Plant membrane sterols: Isolation, identification, and biosynthesis. Meth. Enzymol. 1987, 148, 632–650. [Google Scholar]
- Meance, J.; Duperon, P.; Duperon, R. Localization of sterol compounds inside mitochondria from cauliflower buds. Physiol. Veg. 1976, 14, 745–756. [Google Scholar]
- Yoshida, S.; Uemura, M. Lipid composition of plasma membranes and tonoplasts isolated from etiolated seedlings of mung bean (Vigna radiata L.). Plant Physiol. 1986, 82, 807–812. [Google Scholar] [CrossRef]
- Melkonian, M.; Robenek, H.; Steup, M. Occurrence and distribution of filipin-sterol complexes in chloroplast envelope mem branes of algae and higher plants as visualized by freeze-fracture. Protoplasma 1981, 109, 349–358. [Google Scholar] [CrossRef]
- Li, J.; Nagpal, P.; Vitart, V.; Mcmorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Fujita, S.; Ohnishi, T.; Watanabe, B.; Yokota, T.; Takatsuto, S.; Fujioka, S.; Yoshida, S.; Sakata, K.; Mizutani, M. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 2006, 45, 765–774. [Google Scholar] [CrossRef]
- Men, S.; Boutté, Y.; Ikeda, Y.; Li, X.; Palme, K.; Stierhof, Y.-D.; Hartmann, M.-A.; Moritz, Y.; Grebe, M. Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 2008, 10, 237–244. [Google Scholar] [CrossRef]
- Darnet, S.; Rahier, A. Plant sterol biosynthesis: Identification of two distinct families of sterol 4α-methyl oxidases. Biochem. J. 2004, 378, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, S.; Nie, X.; Boutté, Y.; Grison, M.; Li, P.; Kuang, S.; Men, S. Sterol Methyl Oxidases Affect Embryo Development via Auxin-Associated Mechanisms. Plant Physiol. 2016, 171, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Schaller, H.; Goh, C.; Kwon, M.; Choe, S.; An, C.; Durst, F.; Feldmann, K.; Feyereisen, R. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 2005, 138, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.-C.; Fujioka, S.; Tasaka, M.; Seto, H.; Takatsuto, S.; Isii, A.; Aida, M.; Yoshida, S.; Sheen, J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000, 14, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Schrick, K.; Mayer, U.; Horrichs, A.; Kuhnt, C.; Bellini, C.; Dangl, J.; Schmidt, J.; Jurgens, G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000, 14, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Toopping, J.F.; May, V.J.; Muskett, P.R.; Lindsey, K. Mutations in the HYDRA1 gene of Arabidopsis perturb cell shape and disrupt embryonic and seedling morphogenesis. Development 1997, 124, 4415–4424. [Google Scholar] [CrossRef]
- Schrick, K.; Mayer, U.; Martin, G.; Bellini, C.; Kuhnt, C.; Schmidt, J.; Jürgens, G. Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 2002, 31, 61–73. [Google Scholar] [CrossRef]
- Souter, M.; Topping, J.; Pullen, M.; Friml, J.; Palme, K.; Hackett, R.; Grierson, D.; Lindsey, K. hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 2002, 14, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Diener, A.C.; Li, H.; Zhou, W.; Whoriskey, W.J.; Nes, W.D.; Fink, G.R. Sterol Methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 2000, 12, 853. [Google Scholar] [CrossRef]
- Schaeffer, A.; Bronner, R.; Benveniste, P.; Schaller, H. The ratio of campesterol to sitosterol that modulates growth in Arabidopsis is controlled by STEROL METHYLTRANSFERASE 2;1. Plant J. 2001, 25, 605–615. [Google Scholar] [CrossRef]
- Carland, F.; Fujioka, S.; Nelson, T. The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 2010, 153, 741–756. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Schmit, A.-C.; Heintz, D.; Schaller, H.; Ohta, D. Diversification of sterol methyltransferase enzymes in plants and a role for β-sitosterol in oriented cell plate formation and polarized growth. Plant J. 2015, 84, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, L.J.; Burns, V.; Brown, A.J. A lipidomic perspective on intermediates in cholesterol synthesis as indicators of disease status. J. Genet. Genom. 2014, 41, 275–282. [Google Scholar] [CrossRef]
- Schuler, I.; Duportail, G.; Glasser, N.; Benveniste, P.; Hartmann, M.-A. Soybean phosphatidylcholine vesicles containing plant sterols: A fluorescence anisotropy study. Biochim. Biophys. Acta 1990, 1028, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.-A. Plant sterols and the membrane environment. Trends Plant Sci. 1998, 3, 170–175. [Google Scholar] [CrossRef]
- Schuler, I.; Milon, A.; Nakatani, Y.; Ourisson, G.; Albrecht, A.M.; Benveniste, P.; Hartman, M.-A. Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc. Nat. Acad. Sci. USA 1991, 88, 6926–6930. [Google Scholar] [CrossRef]
- Beck, J.G.; Mathieu, D.; Loudet, C.; Buchoux, S.; Dufourc, E.J. Plant sterols in “rafts”: A better way to regulate membrane thermal shocks. FASEB J. 2007, 21, 1714–1723. [Google Scholar] [CrossRef]
- Dufourc, E.J. Sterols and membrane dynamics. J. Chem. Biol. 2008, 1, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Hodzic, A.; Rappolt, M.; Amenitsch, H.; Laggner, P.; Pabst, G. Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys. J. 2008, 94, 3935–3944. [Google Scholar] [CrossRef] [PubMed]
- Cassim, A.M.; Gouguet, P.; Gronnier, J.; Laurent, N.; Germain, V.; Grison, M.; Boutté, Y.; Gerbeau-Pissot, P.; Simon-Plas, F.; Mongrand, S. Plant lipids: Key players of plasma membrane organization and function. Prog. Lipid Res. 2010, 73, 1–27. [Google Scholar] [CrossRef]
- Grosjean, K.; Mongrand, S.; Beney, L.; Simon-Plas, F.; Gerbeau-Pissot, P. Differential effect of plant lipids on membrane organ ization. J. Biol. Chem. 2015, 290, 5810–5825. [Google Scholar] [CrossRef] [PubMed]
- Sena, F.; Sotelo-Silveira, M.; Astrada, S.; Botella, M.A.; Malacrid, L.; Borsani, O. Spectral phasor analysis reveals altered mem brane order and function of root hair cells in Arabidopsis dry2/sqe1-5 drought hypersensitive mutant. Plant Physiol. Biochem. 2009, 119, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Schaller, H. New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol. Biochem. 2004, 42, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Bouvier-Nave, P.; Husselstein, T.; Benveniste, P. Two families of sterol methyltransferases are involved in the first and the second methylation steps of plant sterol biosynthesis. Eur. J. Biochem. 1998, 256, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, K.; Jones, C.W.; Stephens, C.M.; Vatsyayan, R.; Marshall, J.A.; Nes, W.D. Molecular probing of the Saccharomyces cerevisiae sterol 24-C methyltransferase reveals multiple amino acid residues involved with C2-transfer activity. Biochim. Biophys. Acta 2008, 1781, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Lukowitz, W.; Mayer, U.; Jürgens, G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 1996, 84, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Lauber, M.H.; Waizenegger, I.; Steinmann, T.; Schwarz, H.; Mayer, U.; Hwang, I.; Lukowitz, W.; Jürgens, G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997, 139, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, H.; Liu, P.; Hao, H.; Jin, J.B.; Lin, J. Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS ONE 2011, 6, e26129. [Google Scholar] [CrossRef] [PubMed]
- Feraru, E.; Feraru, M.I.; Asaoka, R.; Paciorek, T.; De Rycke, R.; Tanaka, H.; Nakano, A.; Friml, J. BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 2012, 24, 3074–3086. [Google Scholar] [CrossRef]
- Livanos, P.; Müller, S. Division plane establishment and cytokinesis. Annu. Rev. Plant Biol. 2019, 70, 239–267. [Google Scholar] [CrossRef]
- Ursache, R.; Andersen, T.G.; Marhavý, P.; Geldner, N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018, 93, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Valvekens, D.; Montagu, M.V.; Lusebettens, M.V. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 1998, 85, 5536–5540. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, F.; Yoneda, A.; Tomida, T.; Sano, T.; Nagata, T.; Hasezawa, S. Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: Time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol. 2001, 42, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Enami, K.; Ichikawa, M.; Uemura, T.; Kutsuna, N.; Hasezawa, S.; Nakagawa, T.; Nakano, A.; Sato, M.H. Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, M.; Hirano, T.; Enami, K.; Fuselier, T.; Kato, N.; Kwon, C.; Voigt, B.; Schulze-Lefert, P.; Baluška, F.; Sato, M.H. Syn taxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Fujimoto, M.; Okatani, Y.; Nishiyama, T.; Goh, T.; Ito, E.; Dainobu, T.; Nishitani, A.; Uemura, T.; Sato, M.H.; et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 2001, 13, 853–859. [Google Scholar] [CrossRef]
- Asaoka, R.; Uemura, T.; Ito, J.; Fujimoto, M.; Ito, E.; Ueda, T.; Nakano, A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013, 73, 240–249. [Google Scholar] [CrossRef]
- Sano, T.; Higaki, T.; Oda, Y.; Hayashi, T.; Hasezawa, S. Appearance of actin microfilament ‘twin peaks’ in mitosis and their function in cell plate formation, as visualized in tobacco BY-2 cells expressing GFP-fimbrin. Plant J. 2005, 44, 595–605. [Google Scholar] [CrossRef]
- Kurihara, D.; Mizuta, Y.; Sato, Y.; Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 2015, 142, 4168–4179. [Google Scholar] [CrossRef] [PubMed]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Babiychuk, E.; Bouvier-Nave, P.; Compagnon, V.; Suzuki, M.; Muranaka, T.; Van Montagu, M.; Kushnir, S.; Schaller, H. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3163–3168. [Google Scholar] [CrossRef] [PubMed]
- Bouvier-Navé, P.; Berna, A.; Noiriel, A.; Compagnon, V.; Carlsson, A.S.; Banas, A.; Stymne, S.; Schaller, H. Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 2010, 152, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Fukao, Y.; Nishimura, M.; Hara-Nishimura, I. NAI1 gene encodes a basic-helix-loop-helix–type putative transcription factor that regulates the formation of an endoplasmic reticulum–derived structure, the ER body. Plant Cell 2004, 16, 1536–1549. [Google Scholar] [CrossRef]
- Peng, L.; Kawagoe, Y.; Hogan, P.; Delmer, D. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 2002, 295, 147–150. [Google Scholar] [CrossRef]
- Schrick, K.; DeBolt, S.; Bulone, V. Deciphering the molecular function of sterols in cellulose biosynthesis. Front. Plant Sci. 2012, 3, 84. [Google Scholar] [CrossRef]
- Boutté, Y.; Frescatada-Rosa, M.; Men, S.; Chow, C.-M.; Ebine, K.; Gustavsson, A.; Johansson, L.; Ueda, T.; Moore, I.; Jürgens, G.; et al. Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 2010, 29, 546–558. [Google Scholar] [CrossRef]
- Mahaddalkar, T.; Suri, C.; Naik, P.K.; Lopus, M. Biochemical characterization and molecular dynamic simulation of β-sitosterol as a tubulin-binding anticancer agent. Eur. J. Pharmacol. 2015, 760, 154–162. [Google Scholar] [CrossRef]
- Chen, H.-W.; Persson, S.; Grebe, M.; McFarlane, H.E. Cellulose synthesis during cell plate assembly. Physiol. Pant 2018, 164, 17–26. [Google Scholar] [CrossRef]
- Dubois, G.A.; Jaillais, Y. Anionic phospholipid gradients: An uncharacterized frontier of the plant endomembrane network. Plant Physiol. 2021, 185, 577–592. [Google Scholar] [CrossRef]
- Frescatada-Rosa, M.; Stanislas, T.; Steven, K.; Backues, S.K.; Reichardt, I.; Men, S.; Boutté, Y.; Jürgens, G.; Moritz, T.; Bednarek, S.Y.; et al. High lipid order of Arabidopsis cell-plate membranes mediated by sterol and DYNAMIN-RELATED PROTEIN1A function. Plant J. 2014, 80, 745–757. [Google Scholar] [CrossRef]
- Tang, L.; Li, Y.; Zhong, C.; Deng, X.; Wang, X. Plant sterol clustering correlates with membrane microdomains as revealed by optical and computational microscopy. Membranes 2021, 11, 747. [Google Scholar] [CrossRef]
- Borner, G.H.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; MacAskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef]
- Morel, J.; Claverol, S.; Mongrand, S.; Furt, F.; Fromentin, J.; Bessoule, J.-J.; Blein, J.-P.; Simon-Plas, F. Proteomics of plant deter gent-resistant membranes. Mol. Cell Proteom. 2006, 5, 1396–1411. [Google Scholar] [CrossRef]
- Kierszniowska, S.; Seiwert, B.; Schulze, W.X. Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-β-cyclodextrin and quantitative proteomics. Mol. Cell Proteom. 2009, 8, 612–623. [Google Scholar] [CrossRef]
- Takahashi, D.; Kawamura, Y.; Uemura, M. Detergent-resistant plasma membrane proteome to elucidate microdomain functions in plant cells. Front. Plant Sci. 2013, 4, 27. [Google Scholar] [CrossRef]
- Stanislas, T.; Hüser, A.; Barbosa, I.C.R.; Kiefer, C.S.; Brackmann, K.; Pietra, S.; Gustavsson, A.; Zourelidou, M.; Schwechheimer, C.; Grebe, M. Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat. Plants 2015, 1, 15162. [Google Scholar] [CrossRef]
- Kang, E.; Zheng, M.; Zhang, Y.; Yuan, M.; Yalovsky, S.; Zhu, L.; Fu, Y. The microtubule-associated protein MAP18 affects ROP2 GTPase activity during root hair growth. Plant Physiol. 2017, 174, 202–222. [Google Scholar] [PubMed]
- Smokvarska, M.; Jaillais, Y.; Martinière, A. Function of membrane domains in rho-of-plant signaling. Plant Physiol. 2021, 185, 663–681. [Google Scholar] [PubMed]
- Hollweck, M.; Jordan, D.; Bracher, F. Synthesis of a side chain alkyne analogue of sitosterol as a chemical probe for imaging in plant cells. Biomolecules 2024, 14, 542. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohta, D.; Fuwa, A.; Yamaroku, Y.; Isobe, K.; Nakamoto, M.; Okazawa, A.; Ogawa, T.; Ebine, K.; Ueda, T.; Mercier, P.; et al. Characterization of Subcellular Dynamics of Sterol Methyltransferases Clarifies Defective Cell Division in smt2 smt3, a C-24 Ethyl Sterol-Deficient Mutant of Arabidopsis. Biomolecules 2024, 14, 868. https://doi.org/10.3390/biom14070868
Ohta D, Fuwa A, Yamaroku Y, Isobe K, Nakamoto M, Okazawa A, Ogawa T, Ebine K, Ueda T, Mercier P, et al. Characterization of Subcellular Dynamics of Sterol Methyltransferases Clarifies Defective Cell Division in smt2 smt3, a C-24 Ethyl Sterol-Deficient Mutant of Arabidopsis. Biomolecules. 2024; 14(7):868. https://doi.org/10.3390/biom14070868
Chicago/Turabian StyleOhta, Daisaku, Ayaka Fuwa, Yuka Yamaroku, Kazuki Isobe, Masatoshi Nakamoto, Atsushi Okazawa, Takumi Ogawa, Kazuo Ebine, Takashi Ueda, Pierre Mercier, and et al. 2024. "Characterization of Subcellular Dynamics of Sterol Methyltransferases Clarifies Defective Cell Division in smt2 smt3, a C-24 Ethyl Sterol-Deficient Mutant of Arabidopsis" Biomolecules 14, no. 7: 868. https://doi.org/10.3390/biom14070868
APA StyleOhta, D., Fuwa, A., Yamaroku, Y., Isobe, K., Nakamoto, M., Okazawa, A., Ogawa, T., Ebine, K., Ueda, T., Mercier, P., & Schaller, H. (2024). Characterization of Subcellular Dynamics of Sterol Methyltransferases Clarifies Defective Cell Division in smt2 smt3, a C-24 Ethyl Sterol-Deficient Mutant of Arabidopsis. Biomolecules, 14(7), 868. https://doi.org/10.3390/biom14070868







