Cytokines from SARS-CoV-2 Spike-Activated Macrophages Hinder Proliferation and Cause Cell Dysfunction in Endothelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Experimental Treatments
2.2. Cell Proliferation
2.3. Cell Cycle Analysis
2.4. Cell Death
2.5. Western Blot Analysis
2.6. RT-qPCR Analysis
2.7. Statistical Analysis
2.8. Materials
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pelle, M.C.; Zaffina, I.; Luca, S.; Forte, V.; Trapanese, V.; Melina, M.; Giofre, F.; Arturi, F. Endothelial dysfunction in COVID-19: Potential mechanisms and possible therapeutic options. Life 2022, 12, 1605. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Salazar, B.; Holwerda, M.; Studle, C.; Piragyte, I.; Mercader, N.; Engelhardt, B.; Rieben, R.; Doring, Y. COVID-19 and the vasculature: Current aspects and long-term consequences. Front. Cell Dev. Biol. 2022, 10, 824851. [Google Scholar] [CrossRef] [PubMed]
- Sastry, S.; Cuomo, F.; Muthusamy, J. COVID-19 and thrombosis: The role of hemodynamics. Thromb. Res. 2022, 212, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Staffolani, S.; Iencinella, V.; Cimatti, M.; Tavio, M. Long COVID-19 syndrome as a fourth phase of SARS-CoV-2 infection. Infez. Med. 2022, 30, 22–29. [Google Scholar] [PubMed]
- Batah, S.S.; Fabro, A.T. Pulmonary pathology of ards in COVID-19: A pathological review for clinicians. Respir. Med. 2021, 176, 106239. [Google Scholar] [CrossRef] [PubMed]
- Pu, D.; Zhai, X.; Zhou, Y.; Xie, Y.; Tang, L.; Yin, L.; Liu, H.; Li, L. A narrative review of COVID-19-related acute respiratory distress syndrome (cards): “Typical” or “atypical” ards? Ann. Transl. Med. 2022, 10, 908. [Google Scholar] [CrossRef] [PubMed]
- Empson, S.; Rogers, A.J.; Wilson, J.G. COVID-19 acute respiratory distress syndrome: One pathogen, multiple phenotypes. Crit. Care Clin. 2022, 38, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Zemans, R.L.; Zimmerman, G.A.; Arabi, Y.M.; Beitler, J.R.; Mercat, A.; Herridge, M.; Randolph, A.G.; Calfee, C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 2019, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Rowaiye, A.B.; Okpalefe, O.A.; Onuh Adejoke, O.; Ogidigo, J.O.; Hannah Oladipo, O.; Ogu, A.C.; Oli, A.N.; Olofinase, S.; Onyekwere, O.; Rabiu Abubakar, A.; et al. Attenuating the effects of novel COVID-19 (SARS-CoV-2) infection-induced cytokine storm and the implications. J. Inflamm. Res. 2021, 14, 1487–1510. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of tnf-alpha and ifn-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, P.; Neri, M.; D’Errico, S.; Alfieri, L.; Bonuccelli, D.; Cingolani, M.; Di Paolo, M.; Gaudio, R.M.; Lestani, M.; Marti, M.; et al. Cytokine storm and histopathological findings in 60 cases of COVID-19-related death: From viral load research to immunohistochemical quantification of major players il-1beta, il-6, il-15 and tnf-alpha. Forensic Sci. Med. Pathol. 2022, 18, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Colas-Algora, N.; Munoz-Pinillos, P.; Cacho-Navas, C.; Avendano-Ortiz, J.; de Rivas, G.; Barroso, S.; Lopez-Collazo, E.; Millan, J. Simultaneous targeting of il-1-signaling and il-6-trans-signaling preserves human pulmonary endothelial barrier function during a cytokine storm-brief report. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Long, H.; Sun, J.; Li, H.; He, Y.; Wang, Q.; Pan, K.; Tong, Y.; Wang, B.; Wu, Q.; et al. New laboratory evidence for the association between endothelial dysfunction and COVID-19 disease progression. J. Med. Virol. 2022, 94, 3112–3120. [Google Scholar] [CrossRef] [PubMed]
- Rotoli, B.M.; Barilli, A.; Visigalli, R.; Ferrari, F.; Dall’Asta, V. Endothelial cell activation by SARS-CoV-2 spike s1 protein: A crosstalk between endothelium and innate immune cells. Biomedicines 2021, 9, 1220. [Google Scholar] [CrossRef] [PubMed]
- Ingoglia, F.; Visigalli, R.; Rotoli, B.M.; Barilli, A.; Riccardi, B.; Puccini, P.; Milioli, M.; Di Lascia, M.; Bernuzzi, G.; Dall’Asta, V. Human macrophage differentiation induces octn2-mediated l-carnitine transport through stimulation of mtor-stat3 axis. J. Leukoc. Biol. 2017, 101, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Recchia Luciani, G.; Soli, M.; Dall’Asta, V.; Rotoli, B.M. Growth arrest of alveolar cells in response to cytokines from spike s1-activated macrophages: Role of ifn-gamma. Biomedicines 2022, 10, 3085. [Google Scholar] [CrossRef] [PubMed]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert. Rev. Mol. Med. 2008, 10, e19. [Google Scholar] [CrossRef] [PubMed]
- Giacinti, C.; Giordano, A. Rb and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [PubMed]
- Recchia Luciani, G.; Barilli, A.; Visigalli, R.; Sala, R.; Dall’Asta, V.; Rotoli, B.M. Irf1 mediates growth arrest and the induction of a secretory phenotype in alveolar epithelial cells in response to inflammatory cytokines ifngamma/tnfalpha. Int. J. Mol. Sci. 2024, 25, 3463. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Recchia Luciani, G.; Soli, M.; Dall’Asta, V.; Rotoli, B.M. The jak1/2 inhibitor baricitinib mitigates the spike-induced inflammatory response of immune and endothelial cells in vitro. Biomedicines 2022, 10, 2324. [Google Scholar] [CrossRef] [PubMed]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The nuclear factor kappa b (nf-kb) signaling in cancer development and immune diseases. Genes. Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.W.; Ilyas, I.; Weng, J.P. Endothelial dysfunction in COVID-19: An overview of evidence, biomarkers, mechanisms and potential therapies. Acta Pharmacol. Sin. 2023, 44, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Silaghi-Dumitrescu, R.; Patrascu, I.; Lehene, M.; Bercea, I. Comorbidities of COVID-19 patients. Medicina 2023, 59, 1393. [Google Scholar] [CrossRef] [PubMed]
- Otifi, H.M.; Adiga, B.K. Endothelial dysfunction in COVID-19 infection. Am. J. Med. Sci. 2022, 363, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Lui, K.O.; Ma, Z.; Dimmeler, S. SARS-CoV-2 induced vascular endothelial dysfunction: Direct or indirect effects? Cardiovasc. Res. 2024, 120, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Dri, E.; Lampas, E.; Lazaros, G.; Lazarou, E.; Theofilis, P.; Tsioufis, C.; Tousoulis, D. Inflammatory mediators of endothelial dysfunction. Life 2023, 13, 1420. [Google Scholar] [CrossRef]
- Lyon, C.J.; Law, R.E.; Hsueh, W.A. Minireview: Adiposity, inflammation, and atherogenesis. Endocrinology 2003, 144, 2195–2200. [Google Scholar] [PubMed]
- Zhang, H.; Park, Y.; Wu, J.; Chen, X.; Lee, S.; Yang, J.; Dellsperger, K.C.; Zhang, C. Role of tnf-alpha in vascular dysfunction. Clin. Sci. 2009, 116, 219–230. [Google Scholar]
- Ng, C.T.; Fong, L.Y.; Abdullah, M.N.H. Interferon-gamma (ifn-gamma): Reviewing its mechanisms and signaling pathways on the regulation of endothelial barrier function. Cytokine 2023, 166, 156208. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Kang, K.W.; Seu, Y.B.; Baek, S.H.; Kim, J.R. Interferon-gamma induces cellular senescence through p53-dependent DNA damage signaling in human endothelial cells. Mech. Ageing Dev. 2009, 130, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Cantini, G.; Mello, T.; Francalanci, M.; Gelmini, S.; Cosmi, L.; Santarlasci, V.; Degl’Innocenti, S.; Luciani, P.; Deledda, C.; et al. Molecular mechanisms underlying the pro-inflammatory synergistic effect of tumor necrosis factor alpha and interferon gamma in human microvascular endothelium. Eur. J. Cell Biol. 2009, 88, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.N.; Teague, H.L.; Swindell, W.R.; Baumer, Y.; Ward, N.L.; Xing, X.; Baugous, B.; Johnston, A.; Joshi, A.A.; Silverman, J.; et al. Ifn-gamma and tnf-alpha synergism may provide a link between psoriasis and inflammatory atherogenesis. Sci. Rep. 2017, 7, 13831. [Google Scholar] [CrossRef]
- Kandhaya-Pillai, R.; Yang, X.; Tchkonia, T.; Martin, G.M.; Kirkland, J.L.; Oshima, J. Tnf-alpha/ifn-gamma synergy amplifies senescence-associated inflammation and SARS-CoV-2 receptor expression via hyper-activated jak/stat1. Aging Cell 2022, 21, e13646. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, S.; Zhao, L.; Zhang, B.; Chen, H. Ifn-gamma and tnf-alpha aggravate endothelial damage caused by cd123-targeted car t cell. Onco Targets Ther. 2019, 12, 4907–4925. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Serrano, A.; Salero, E.; Tovar, A.; Amescua, G.; Galor, A.; Keane, R.W.; de Rivero Vaccari, J.P.; Sabater, A.L. Tumor necrosis factor-alpha and interferon-gamma induce inflammasome-mediated corneal endothelial cell death. Exp. Eye Res. 2021, 207, 108574. [Google Scholar] [CrossRef]
- Moser, J.; Miller, I.; Carter, D.; Spencer, S.L. Control of the restriction point by rb and p21. Proc. Natl. Acad. Sci. USA 2018, 115, E8219–E8227. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Recchia Luciani, G.; Visigalli, R.; Sala, R.; Soli, M.; Dall’Asta, V.; Rotoli, B.M. Cytokine-induced inos in a549 alveolar epithelial cells: A potential role in COVID-19 lung pathology. Biomedicines 2023, 11, 2699. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, Y.; Schreiber, R.D.; Hamilton, T.A. Synergy between interferon-gamma and tumor necrosis factor-alpha in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor kappab. J. Biol. Chem. 1997, 272, 14899–14907. [Google Scholar] [CrossRef] [PubMed]
- Miki, H.; Ohmori, Y. Transcriptional synergism between nf-κb and stat1. J. Oral Biosci. 2005, 47, 230–242. [Google Scholar]
- Wang, C.; Qin, L.; Manes, T.D.; Kirkiles-Smith, N.C.; Tellides, G.; Pober, J.S. Rapamycin antagonizes tnf induction of vcam-1 on endothelial cells by inhibiting mtorc2. J. Exp. Med. 2014, 211, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, D.; Ernandez, T.; Rosetti, F.; Batal, I.; Cullere, X.; Luscinskas, F.W.; Zhang, Y.; Stavrakis, G.; Garcia-Cardena, G.; Horwitz, B.H.; et al. Endothelial tnf receptor 2 induces irf1 transcription factor-dependent interferon-beta autocrine signaling to promote monocyte recruitment. Immunity 2013, 38, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
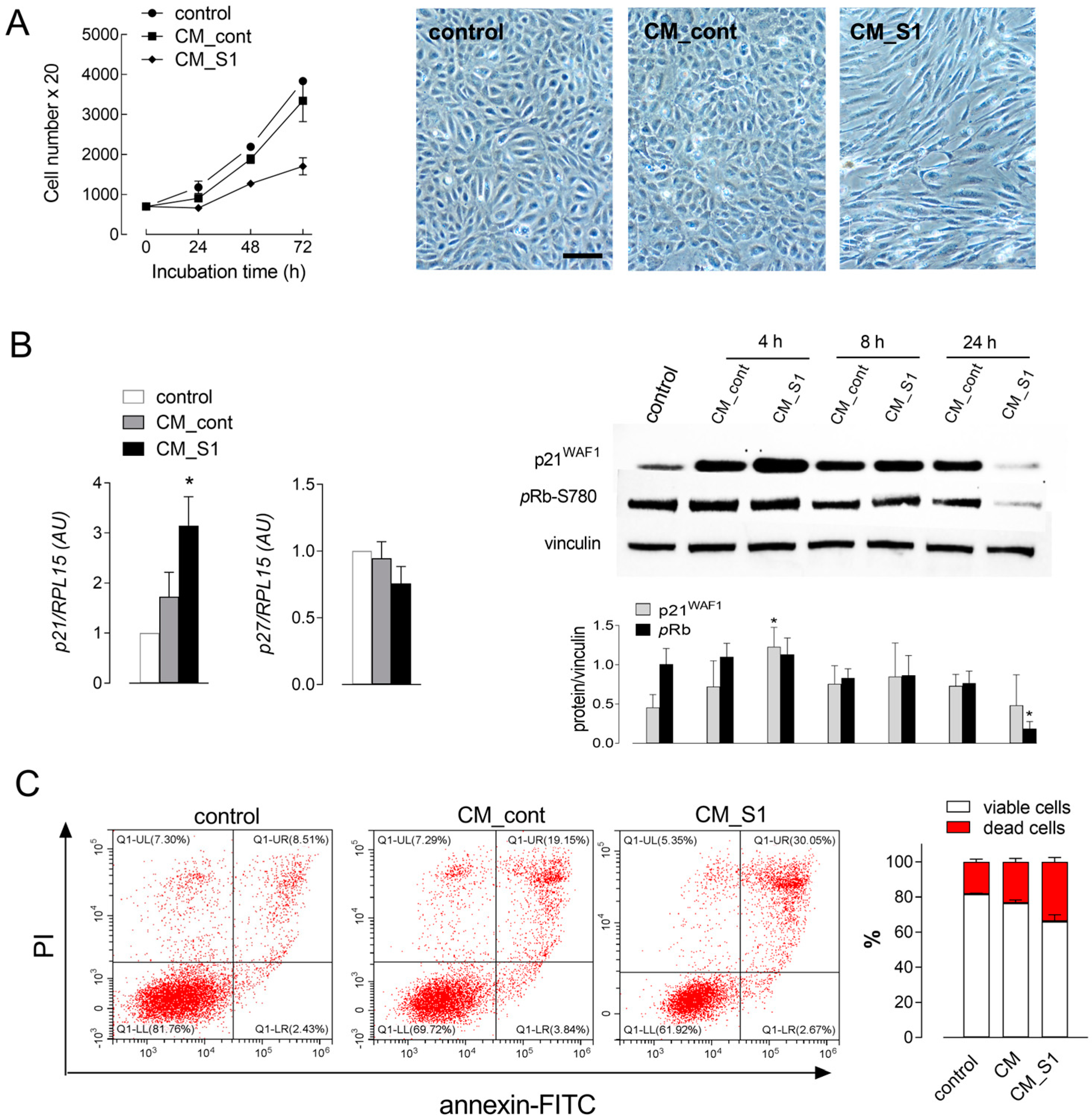
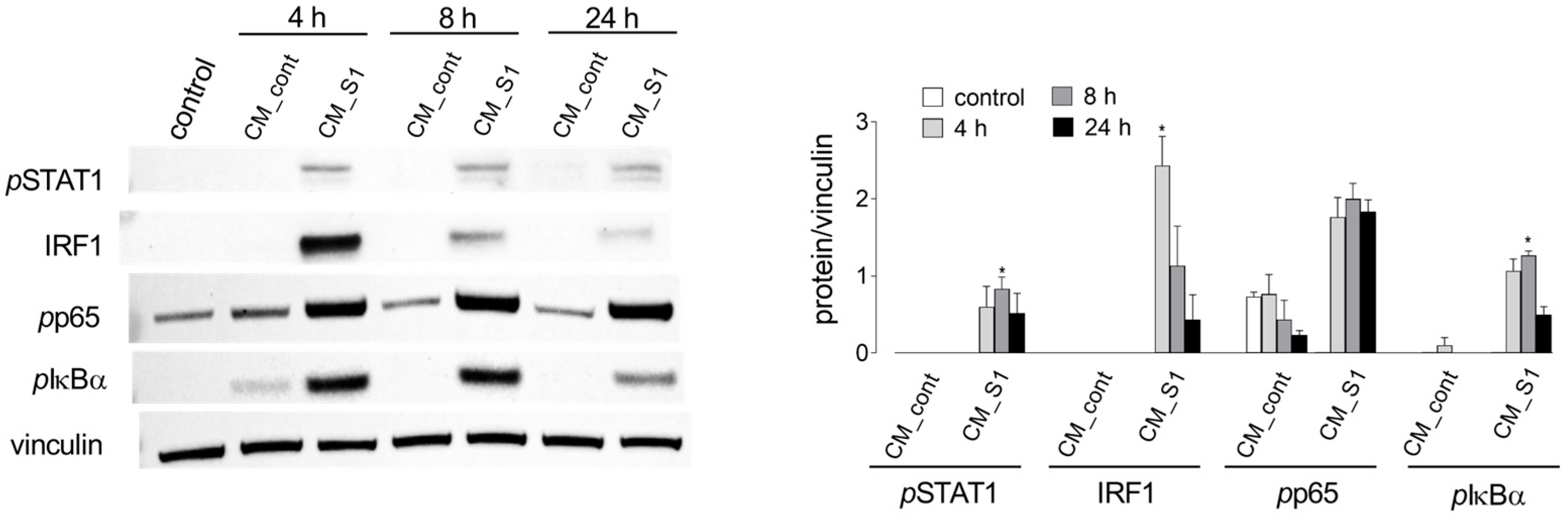
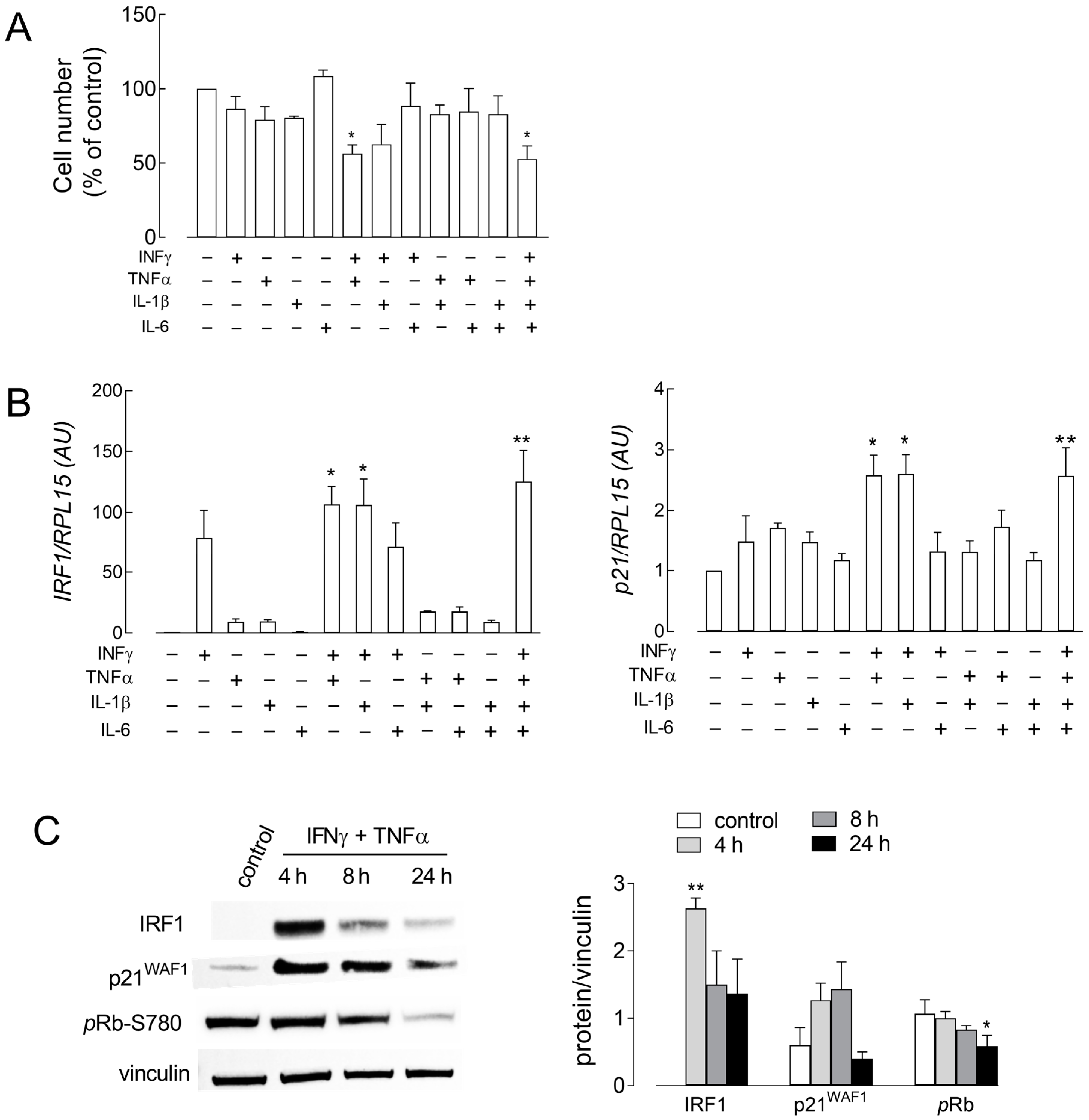
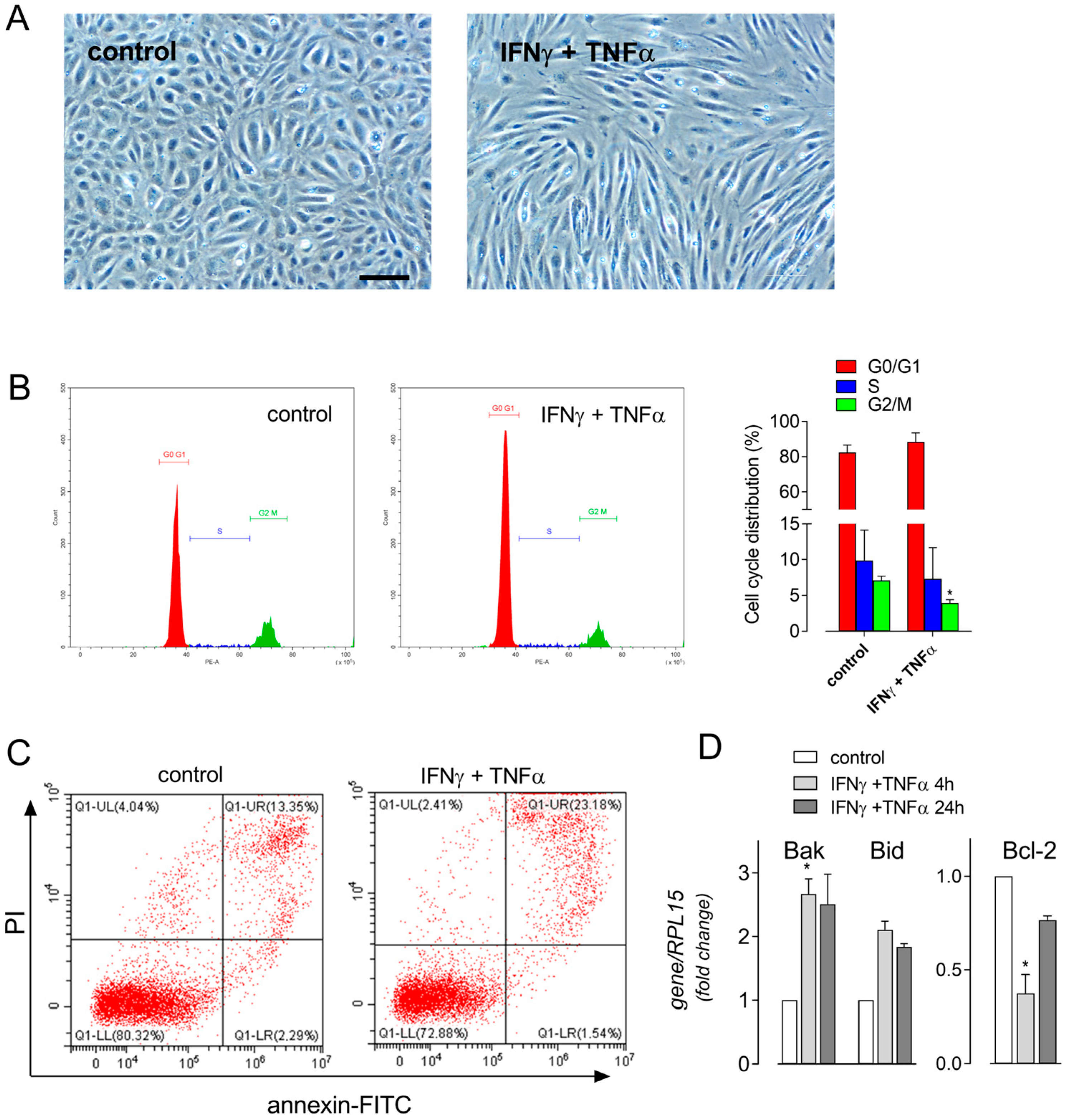
| Gene/Protein Name (Gene ID) | Forward Primer | Reverse Primer |
|---|---|---|
| BAK1/BAK (578) | GAGATGGTCACCTTACCTCTGC | TCATAGCGTCGGTTGATGTCG |
| BCL2/Bcl-2 (596) | ATCGCCCTGTGGATGACTGAGT | GCCAGGAGAAATCAAACAGAGGC |
| BID/BID (637) | ATGGACCGTAGCATCCCTCC | GTAGGTGCGTAGGTTCTGGT |
| CDKN1A/p21 (1026) | CCTGTCACTGTCTTGTACCCT | GCGTTTGGAGTGGTAGAAATCT |
| CDKN1B/p27 (1027) | TAATTGGGGCTCCGGCTAACT | TGCAGGTCGCTTCCTTATTCC |
| IRF1/IRF1 (3659) | CTGTGCGAGTGTACCGGATG | ATCCCCACATGACTTCCTCTT |
| RPL15/RPL15 (6138) | GCAGCCATCAGGTAAGCCAAG | AGCGGACCCTCAGAAGAAAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recchia Luciani, G.; Barilli, A.; Visigalli, R.; Dall’Asta, V.; Rotoli, B.M. Cytokines from SARS-CoV-2 Spike-Activated Macrophages Hinder Proliferation and Cause Cell Dysfunction in Endothelial Cells. Biomolecules 2024, 14, 927. https://doi.org/10.3390/biom14080927
Recchia Luciani G, Barilli A, Visigalli R, Dall’Asta V, Rotoli BM. Cytokines from SARS-CoV-2 Spike-Activated Macrophages Hinder Proliferation and Cause Cell Dysfunction in Endothelial Cells. Biomolecules. 2024; 14(8):927. https://doi.org/10.3390/biom14080927
Chicago/Turabian StyleRecchia Luciani, Giulia, Amelia Barilli, Rossana Visigalli, Valeria Dall’Asta, and Bianca Maria Rotoli. 2024. "Cytokines from SARS-CoV-2 Spike-Activated Macrophages Hinder Proliferation and Cause Cell Dysfunction in Endothelial Cells" Biomolecules 14, no. 8: 927. https://doi.org/10.3390/biom14080927
APA StyleRecchia Luciani, G., Barilli, A., Visigalli, R., Dall’Asta, V., & Rotoli, B. M. (2024). Cytokines from SARS-CoV-2 Spike-Activated Macrophages Hinder Proliferation and Cause Cell Dysfunction in Endothelial Cells. Biomolecules, 14(8), 927. https://doi.org/10.3390/biom14080927







