Radiosensitizing Effect of PARP Inhibition on Chondrosarcoma and Chondrocyte Cells Is Dependent on Radiation LET
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Drug Treatments
2.3. Irradiation
2.4. Clonogenic Assay
2.5. MTT Toxicity Test
2.6. Sequencing
2.7. Western Blotting Analysis
2.8. Statistics
3. Results
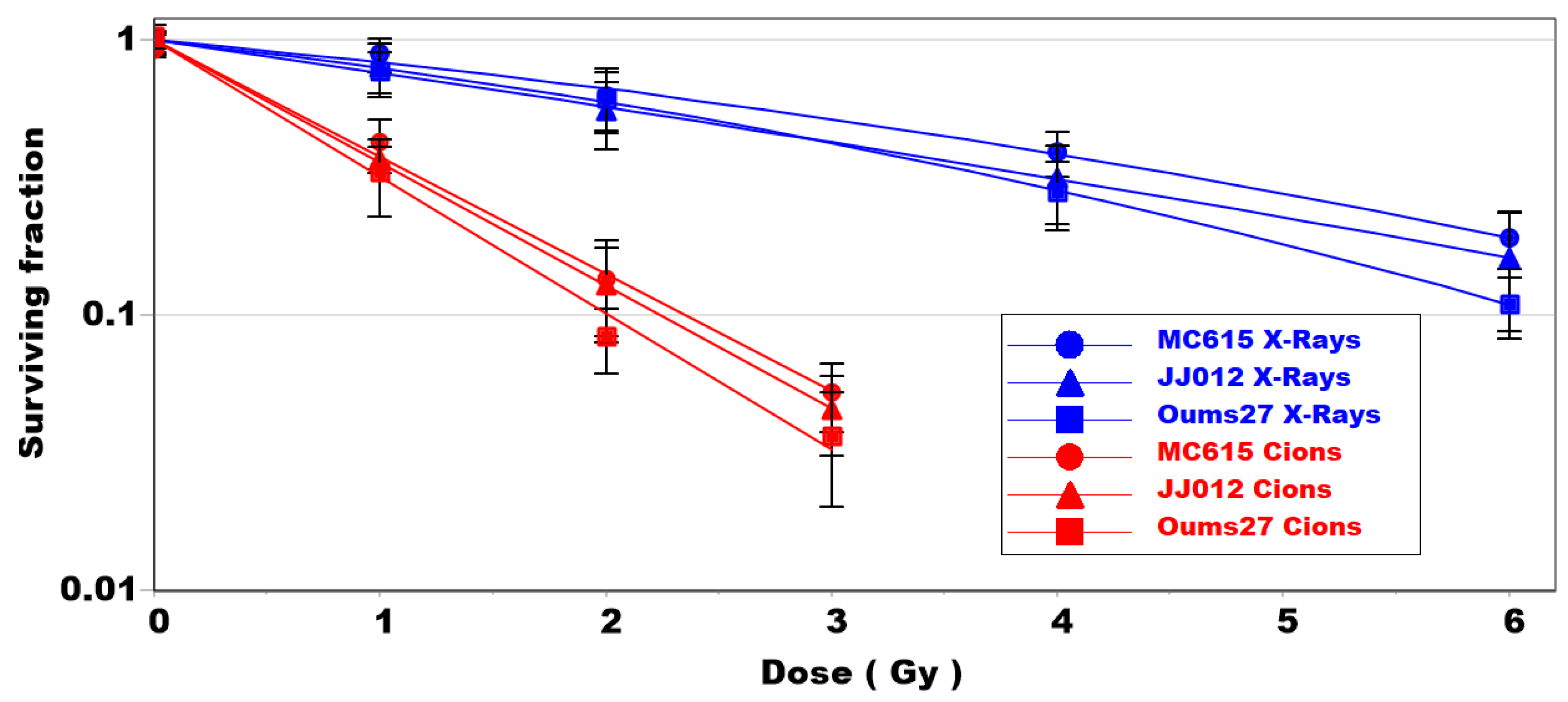
3.1. Clonogenic Survival of Chondrosarcoma and Chondrocyte Cells Is Dependent on LET
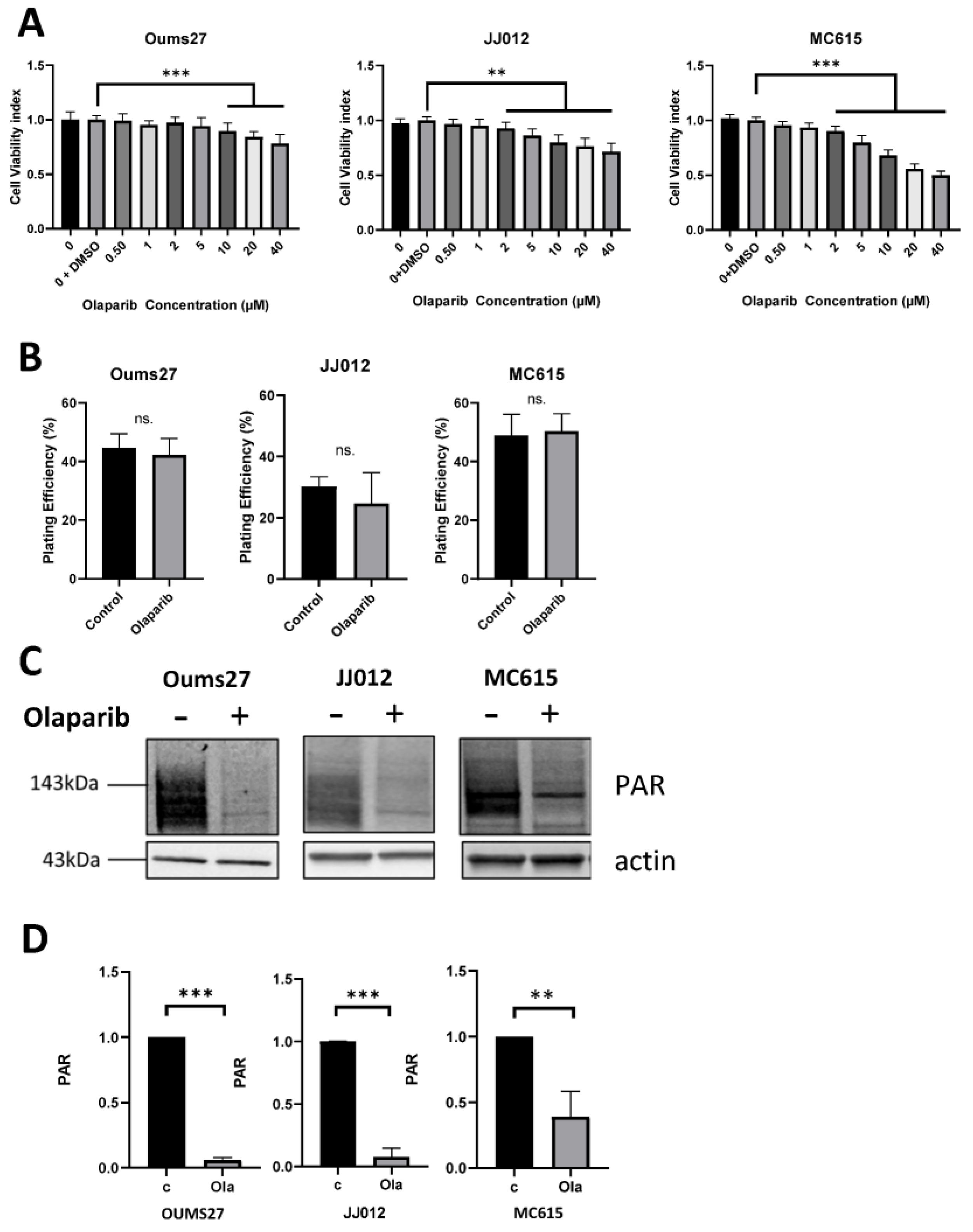
3.2. Similar Low Toxicity and Effective Activity of PARP Inhibitor
3.3. Differential Mutation Status and Genomic Instability: GIScar Score
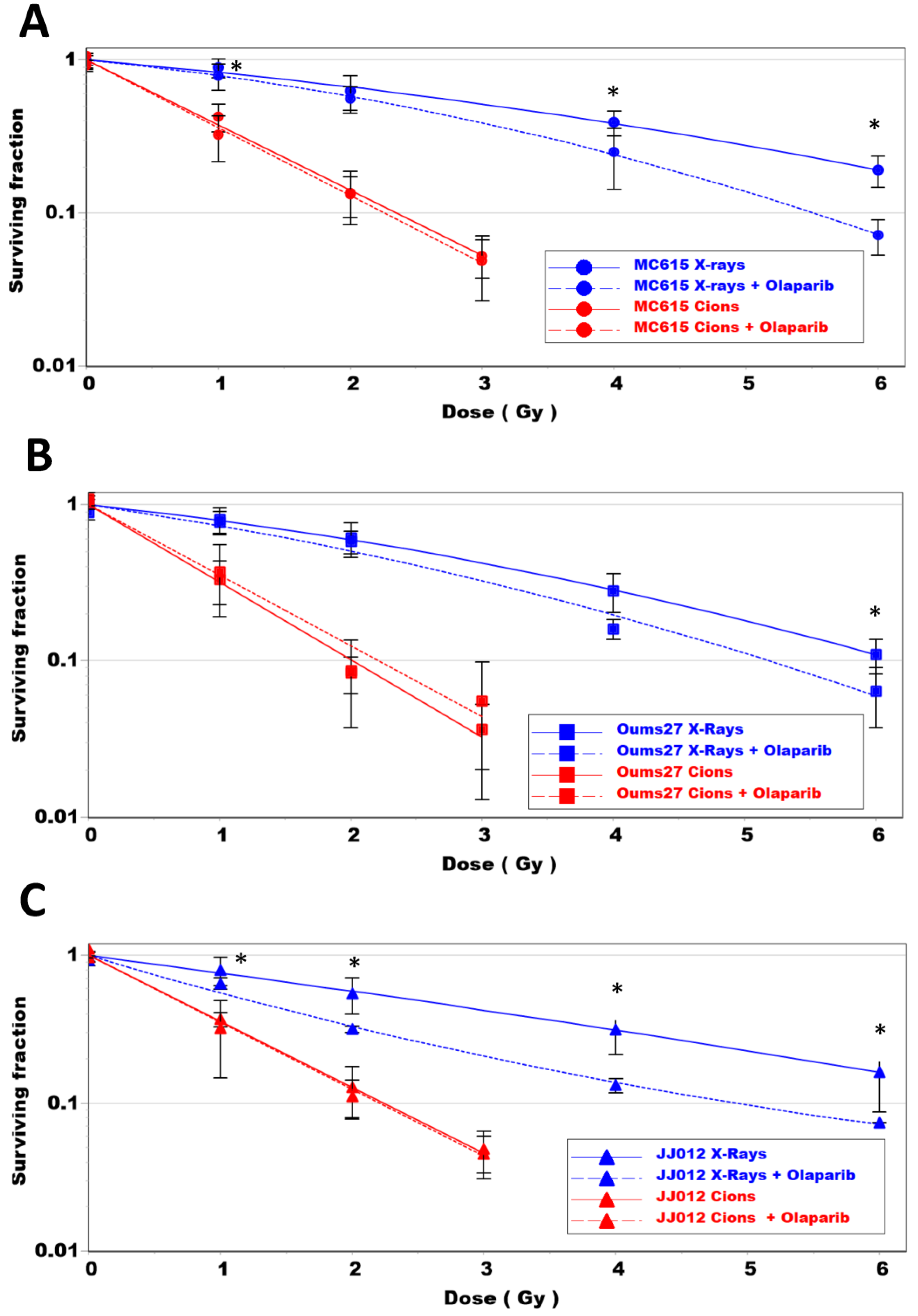
3.4. Radiosensitizing Effect of PARPi Depends on Radiation LET
3.5. Specific Activation of DNA Repair Pathways Depends on Radiation LET
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rozeman, L.B.; Hogendoorn, P.C.W.; Bovée, J.V.M.G. Diagnosis and Prognosis of Chondrosarcoma of Bone. Expert Rev. Mol. Diagn. 2002, 2, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Moussavi-Harami, F.; Mollano, A.; Martin, J.A.; Ayoob, A.; Domann, F.E.; Gitelis, S.; Buckwalter, J.A. Intrinsic Radiation Resistance in Human Chondrosarcoma Cells. Biochem. Biophys. Res. Commun. 2006, 346, 379–385. [Google Scholar] [CrossRef]
- Gelderblom, H.; Hogendoorn, P.C.W.; Dijkstra, S.D.; van Rijswijk, C.S.; Krol, A.D.; Taminiau, A.H.M.; Bovée, J.V.M.G. The Clinical Approach Towards Chondrosarcoma. Oncologist 2008, 13, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Ma, W.; He, X.; Jha, R.K. Review of Therapeutic Strategies for Osteosarcoma, Chondrosarcoma, and Ewing’s Sarcoma. Med. Sci. Monit. 2011, 17, RA177–RA190. [Google Scholar] [CrossRef]
- Maruyama, K.; Imai, R.; Kamada, T.; Tsuji, H.; Tsujii, H. Carbon Ion Radiation Therapy for Chondrosarcoma. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, S139. [Google Scholar] [CrossRef]
- Imai, R.; Kamada, T.; Araki, N.; Working Group for Bone and Soft-Tissue Sarcomas. Clinical Efficacy of Carbon Ion Radiotherapy for Unresectable Chondrosarcomas. Anticancer Res. 2017, 37, 6959–6964. [Google Scholar] [CrossRef] [PubMed]
- Monga, V.; Mani, H.; Hirbe, A.; Milhem, M. Non-Conventional Treatments for Conventional Chondrosarcoma. Cancers 2020, 12, 1962. [Google Scholar] [CrossRef]
- Riva, G.; Cavallo, I.; Gandini, S.; Ingargiola, R.; Pecorilla, M.; Imparato, S.; Rossi, E.; Mirandola, A.; Ciocca, M.; Orlandi, E.; et al. Particle Radiotherapy for Skull Base Chondrosarcoma: A Clinical Series from Italian National Center for Oncological Hadrontherapy. Cancers 2021, 13, 4423. [Google Scholar] [CrossRef]
- Zając, A.E.; Kopeć, S.; Szostakowski, B.; Spałek, M.J.; Fiedorowicz, M.; Bylina, E.; Filipowicz, P.; Szumera-Ciećkiewicz, A.; Tysarowski, A.; Czarnecka, A.M.; et al. Chondrosarcoma-from Molecular Pathology to Novel Therapies. Cancers 2021, 13, 2390. [Google Scholar] [CrossRef]
- Lesueur, P.; Chevalier, F.; Austry, J.-B.; Waissi, W.; Burckel, H.; Noël, G.; Habrand, J.-L.; Saintigny, Y.; Joly, F. Poly-(ADP-Ribose)-Polymerase Inhibitors as Radiosensitizers: A Systematic Review of Pre-Clinical and Clinical Human Studies. Oncotarget 2017, 8, 69105–69124. [Google Scholar] [CrossRef]
- Césaire, M.; Thariat, J.; Candéias, S.M.; Stefan, D.; Saintigny, Y.; Chevalier, F. Combining PARP Inhibition, Radiation, and Immunotherapy: A Possible Strategy to Improve the Treatment of Cancer? Int. J. Mol. Sci. 2018, 19, 3793. [Google Scholar] [CrossRef]
- Lesueur, P.; Chevalier, F.; El-Habr, E.A.; Junier, M.-P.; Chneiweiss, H.; Castera, L.; Müller, E.; Stefan, D.; Saintigny, Y. Radiosensitization Effect of Talazoparib, a Parp Inhibitor, on Glioblastoma Stem Cells Exposed to Low and High Linear Energy Transfer Radiation. Sci. Rep. 2018, 8, 3664. [Google Scholar] [CrossRef] [PubMed]
- Césaire, M.; Ghosh, U.; Austry, J.-B.; Muller, E.; Cammarata, F.P.; Guillamin, M.; Caruso, M.; Castéra, L.; Petringa, G.; Cirrone, G.A.P.; et al. Sensitization of Chondrosarcoma Cells with PARP Inhibitor and High-LET Radiation. J. Bone Oncol. 2019, 17, 100246. [Google Scholar] [CrossRef] [PubMed]
- Thariat, J.; Valable, S.; Laurent, C.; Haghdoost, S.; Pérès, E.A.; Bernaudin, M.; Sichel, F.; Lesueur, P.; Césaire, M.; Petit, E.; et al. Hadrontherapy Interactions in Molecular and Cellular Biology. Int. J. Mol. Sci. 2020, 21, 133. [Google Scholar] [CrossRef] [PubMed]
- Rødland, G.E.; Temelie, M.; Eek Mariampillai, A.; Hauge, S.; Gilbert, A.; Chevalier, F.; Savu, D.I.; Syljuåsen, R.G. Potential Benefits of Combining Proton or Carbon Ion Therapy with DNA Damage Repair Inhibitors. Cells 2024, 13, 1058. [Google Scholar] [CrossRef]
- Brown, J.S.; O’Carrigan, B.; Jackson, S.P.; Yap, T.A. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2017, 7, 20–37. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Montaldi, A.P.; Lima, S.C.G.; Godoy, P.R.D.V.; Xavier, D.J.; Sakamoto-Hojo, E.T. PARP-1 Inhibition Sensitizes Temozolomide-treated Glioblastoma Cell Lines and Decreases Drug Resistance Independent of MGMT Activity and PTEN Proficiency. Oncol. Rep. 2020, 44, 2275–2287. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Radivoyevitch, T.; Nagata, Y.; Khurshed, M.; Przychodzen, B.; Makishima, H.; Xu, M.; Bleeker, F.E.; Wilmink, J.W.; Carraway, H.E.; et al. IDH1/2 Mutations Sensitize Acute Myeloid Leukemia to PARP Inhibition and This Is Reversed by IDH1/2-Mutant Inhibitors. Clin. Cancer Res. 2018, 24, 1705–1715. [Google Scholar] [CrossRef]
- Eder, J.P.; Doroshow, D.B.; Do, K.T.; Keedy, V.L.; Sklar, J.S.; Glazer, P.; Bindra, R.; Shapiro, G.I. Clinical Efficacy of Olaparib in IDH1/IDH2-Mutant Mesenchymal Sarcomas. JCO Precis. Oncol. 2021, 5, 466–472. [Google Scholar] [CrossRef]
- Venneker, S.; Kruisselbrink, A.B.; Briaire-de Bruijn, I.H.; de Jong, Y.; van Wijnen, A.J.; Danen, E.H.J.; Bovée, J.V.M.G. Inhibition of PARP Sensitizes Chondrosarcoma Cell Lines to Chemo- and Radiotherapy Irrespective of the IDH1 or IDH2 Mutation Status. Cancers 2019, 11, 1918. [Google Scholar] [CrossRef] [PubMed]
- Palubeckaitė, I.; Venneker, S.; Briaire-de Bruijn, I.H.; van den Akker, B.E.; Krol, A.D.; Gelderblom, H.; Bovée, J.V.M.G. Selection of Effective Therapies Using Three-Dimensional In Vitro Modeling of Chondrosarcoma. Front. Mol. Biosci. 2020, 7, 566291. [Google Scholar] [CrossRef]
- Kunisada, T.; Miyazaki, M.; Mihara, K.; Gao, C.; Kawai, A.; Inoue, H.; Namba, M. A New Human Chondrosarcoma Cell Line (OUMS-27) That Maintains Chondrocytic Differentiation. Int. J. Cancer 1998, 77, 854–859. [Google Scholar] [CrossRef]
- Scully, S.P.; Berend, K.R.; Toth, A.; Qi, W.N.; Qi, Z.; Block, J.A. Marshall Urist Award. Interstitial Collagenase Gene Expression Correlates with In Vitro Invasion in Human Chondrosarcoma. Clin. Orthop. Relat. Res. 2000, 376, 291–303. [Google Scholar] [CrossRef]
- Mallein-Gerin, F.; Olsen, B.R. Expression of Simian Virus 40 Large T (Tumor) Oncogene in Mouse Chondrocytes Induces Cell Proliferation without Loss of the Differentiated Phenotype. Proc. Natl. Acad. Sci. USA 1993, 90, 3289–3293. [Google Scholar] [CrossRef]
- Valcourt, U.; Gouttenoire, J.; Moustakas, A.; Herbage, D.; Mallein-Gerin, F. Functions of Transforming Growth Factor-β Family Type I Receptors and Smad Proteins in the Hypertrophic Maturation and Osteoblastic Differentiation of Chondrocytes. J. Biol. Chem. 2002, 277, 33545–33558. [Google Scholar] [CrossRef] [PubMed]
- Ronzière, M.-C.; Aubert-Foucher, E.; Gouttenoire, J.; Bernaud, J.; Herbage, D.; Mallein-Gerin, F. Integrin Alpha1beta1 Mediates Collagen Induction of MMP-13 Expression in MC615 Chondrocytes. Biochim. Biophys. Acta 2005, 1746, 55–64. [Google Scholar] [CrossRef]
- Ulivi, V.; Giannoni, P.; Gentili, C.; Cancedda, R.; Descalzi, F. P38/NF-KB-Dependent Expression of COX-2 during Differentiation and Inflammatory Response of Chondrocytes. J. Cell. Biochem. 2008, 104, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Durantel, F.; Balanzat, E.; Cassimi, A.; Chevalier, F.; Ngono-Ravache, Y.; Madi, T.; Poully, J.-C.; Ramillon, J.-M.; Rothard, H.; Ropars, F.; et al. Dosimetry for Radiobiology Experiments at GANIL. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2016, 816, 70–77. [Google Scholar] [CrossRef]
- CS-Cal: A Dedicated Tool for Evaluation of Clonogenic Assays. Available online: https://angiogenesis.dkfz.de/oncoexpress/software/cs-cal/ (accessed on 11 July 2024).
- Lepleux, C.; Marie-Brasset, A.; Temelie, M.; Boulanger, M.; Brotin, É.; Goldring, M.B.; Hirtz, C.; Varès, G.; Nakajima, T.; Saintigny, Y.; et al. Bystander Effectors of Chondrosarcoma Cells Irradiated at Different LET Impair Proliferation of Chondrocytes. J. Cell Commun. Signal. 2019, 13, 343–356. [Google Scholar] [CrossRef]
- Leman, R.; Muller, E.; Legros, A.; Goardon, N.; Chentli, I.; Atkinson, A.; Tranchant, A.; Castera, L.; Krieger, S.; Ricou, A.; et al. Validation of the Clinical Use of GIScar, an Academic-Developed Genomic Instability Score Predicting Sensitivity to Maintenance Olaparib for Ovarian Cancer. Clin. Cancer Res. 2023, 29, 4419–4429. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Goardon, N.; Brault, B.; Rousselin, A.; Paimparay, G.; Legros, A.; Fouillet, R.; Bruet, O.; Tranchant, A.; Domin, F.; et al. OutLyzer: Software for Extracting Low-Allele-Frequency Tumor Mutations from Sequencing Background Noise in Clinical Practice. Oncotarget 2016, 7, 79485–79493. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Groupe Génétique et Cancer. Available online: https://recherche.unicancer.fr/fr/les-groupes-d-experts/groupe-genetique-et-cancer/ (accessed on 25 April 2024).
- Gilbert, A.; Tudor, M.; Montanari, J.; Commenchail, K.; Savu, D.I.; Lesueur, P.; Chevalier, F. Chondrosarcoma Resistance to Radiation Therapy: Origins and Potential Therapeutic Solutions. Cancers 2023, 15, 1962. [Google Scholar] [CrossRef]
- Kim, H.; Cho, Y.; Kim, H.-S.; Kang, D.; Cheon, D.; Kim, Y.-J.; Chang, M.J.; Lee, K.M.; Chang, C.B.; Kang, S.-B.; et al. A System-Level Approach Identifies HIF-2α as a Critical Regulator of Chondrosarcoma Progression. Nat. Commun. 2020, 11, 5023. [Google Scholar] [CrossRef]
- Hu, X.; Li, L.; Eid, J.E.; Liu, C.; Yu, J.; Yue, J.; Trent, J.C. IDH1 Mutation Induces HIF-1α and Confers Angiogenic Properties in Chondrosarcoma JJ012 Cells. Dis. Markers 2022, 2022, 7729968. [Google Scholar] [CrossRef]
- Tlemsani, C.; Larousserie, F.; De Percin, S.; Audard, V.; Hadjadj, D.; Chen, J.; Biau, D.; Anract, P.; Terris, B.; Goldwasser, F.; et al. Biology and Management of High-Grade Chondrosarcoma: An Update on Targets and Treatment Options. Int. J. Mol. Sci. 2023, 24, 1361. [Google Scholar] [CrossRef]
- Chevalier, F.; Hamdi, D.H.; Lepleux, C.; Temelie, M.; Nicol, A.; Austry, J.B.; Lesueur, P.; Vares, G.; Savu, D.; Nakajima, T.; et al. High LET Radiation Overcomes In Vitro Resistance to X-Rays of Chondrosarcoma Cell Lines. Technol. Cancer Res. Treat 2019, 18, 1533033819871309. [Google Scholar] [CrossRef]
- Ghorai, A.; Sarma, A.; Chowdhury, P.; Ghosh, U. PARP-1 Depletion in Combination with Carbon Ion Exposure Significantly Reduces MMPs Activity and Overall Increases TIMPs Expression in Cultured HeLa Cells. Radiat. Oncol. 2016, 11, 126. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhao, M.; Bai, M.; Xue, Z.; Wang, F.; Zhu, Z.; Yu, J.; Yue, J. PARP Inhibitor plus Radiotherapy Reshape the Immune Suppressive Microenvironment and Potentiate the Efficacy of Immune Checkpoint Inhibitors in Tumors with IDH1 Mutation. Cancer Lett. 2024, 586, 216676. [Google Scholar] [CrossRef]
- Tinganelli, W.; Durante, M. Carbon Ion Radiobiology. Cancers 2020, 12, 3022. [Google Scholar] [CrossRef]
- Park, S.; Choi, C.; Kim, H.; Shin, Y.J.; Oh, Y.; Park, W.; Cho, W.K.; Kim, N. Olaparib Enhances Sensitization of BRCA-Proficient Breast Cancer Cells to x-Rays and Protons. Breast Cancer Res. Treat. 2024, 203, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]


| D10 | RBE | ER | ||
|---|---|---|---|---|
| OUMS27 | X-ray | 6.17 | ||
| X-ray + Olaparib | 5.18 | 1.19 * | ||
| C-ions | 2.02 | 3.05 | ||
| C-ions + Olaparib | 2.22 | 0.91 | ||
| JJ012 | X-ray | 7.5 | ||
| X-ray + Olaparib | 5.04 | 1.5 * | ||
| C-ions | 2.24 | 3.37 | ||
| C-ions + Olaparib | 2.22 | 1.01 | ||
| MC615 | X-ray | 7.58 | ||
| X-ray + Olaparib | 5.52 | 1.37 * | ||
| C-ions | 2.36 | 3.21 | ||
| C-ions + Olaparib | 2.27 | 1.04 |
| Cell Line | Gene | Event | Mutation | GIScar Score | Number of Large Genomic Events * |
|---|---|---|---|---|---|
| JJ012 | IDH1 | missense substitution | p.Arg132Gly | ||
| SLX4 | missense substitution | p.Thr345Asn | |||
| TP53 | missense substitution | p.Gly199Val | 6 | ||
| ARID1A | frameshift | p.Gln1552Hisfs*21 | 0.154 | ||
| ARID1B | missense substitution | p.Pro1019Leu | |||
| ARID1B | frameshift | p.Gly351Metfs*11 | |||
| OUMS27 | TP53 | frameshift | p.Ser149Tyrfs*31 | 0.408 | 27 |
| EMSY | intronic mutation | g.76260993C>A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilbert, A.; Tudor, M.; Delaunay, A.; Leman, R.; Levilly, J.; Atkinson, A.; Castéra, L.; Dinischiotu, A.; Savu, D.I.; Valable, S.; et al. Radiosensitizing Effect of PARP Inhibition on Chondrosarcoma and Chondrocyte Cells Is Dependent on Radiation LET. Biomolecules 2024, 14, 1071. https://doi.org/10.3390/biom14091071
Gilbert A, Tudor M, Delaunay A, Leman R, Levilly J, Atkinson A, Castéra L, Dinischiotu A, Savu DI, Valable S, et al. Radiosensitizing Effect of PARP Inhibition on Chondrosarcoma and Chondrocyte Cells Is Dependent on Radiation LET. Biomolecules. 2024; 14(9):1071. https://doi.org/10.3390/biom14091071
Chicago/Turabian StyleGilbert, Antoine, Mihaela Tudor, Amandine Delaunay, Raphaël Leman, Julien Levilly, Alexandre Atkinson, Laurent Castéra, Anca Dinischiotu, Diana Iulia Savu, Samuel Valable, and et al. 2024. "Radiosensitizing Effect of PARP Inhibition on Chondrosarcoma and Chondrocyte Cells Is Dependent on Radiation LET" Biomolecules 14, no. 9: 1071. https://doi.org/10.3390/biom14091071
APA StyleGilbert, A., Tudor, M., Delaunay, A., Leman, R., Levilly, J., Atkinson, A., Castéra, L., Dinischiotu, A., Savu, D. I., Valable, S., & Chevalier, F. (2024). Radiosensitizing Effect of PARP Inhibition on Chondrosarcoma and Chondrocyte Cells Is Dependent on Radiation LET. Biomolecules, 14(9), 1071. https://doi.org/10.3390/biom14091071







