Proteomic Profile of Circulating Extracellular Vesicles in the Brain after Δ9-Tetrahydrocannabinol Inhalation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Drugs
2.3. Experimental Design
2.3.1. Primary Choroid Plexus Epithelial Cell Culture
2.3.2. Targeted RNA Expression Analysis
2.3.3. Passive Vapor THC Administration
2.3.4. CSF Collection and EV Extraction from Rats
2.3.5. Label-Free Quantitative Proteomics Analyses via High-Resolution Tandem Mass Spectrometry
3. Results
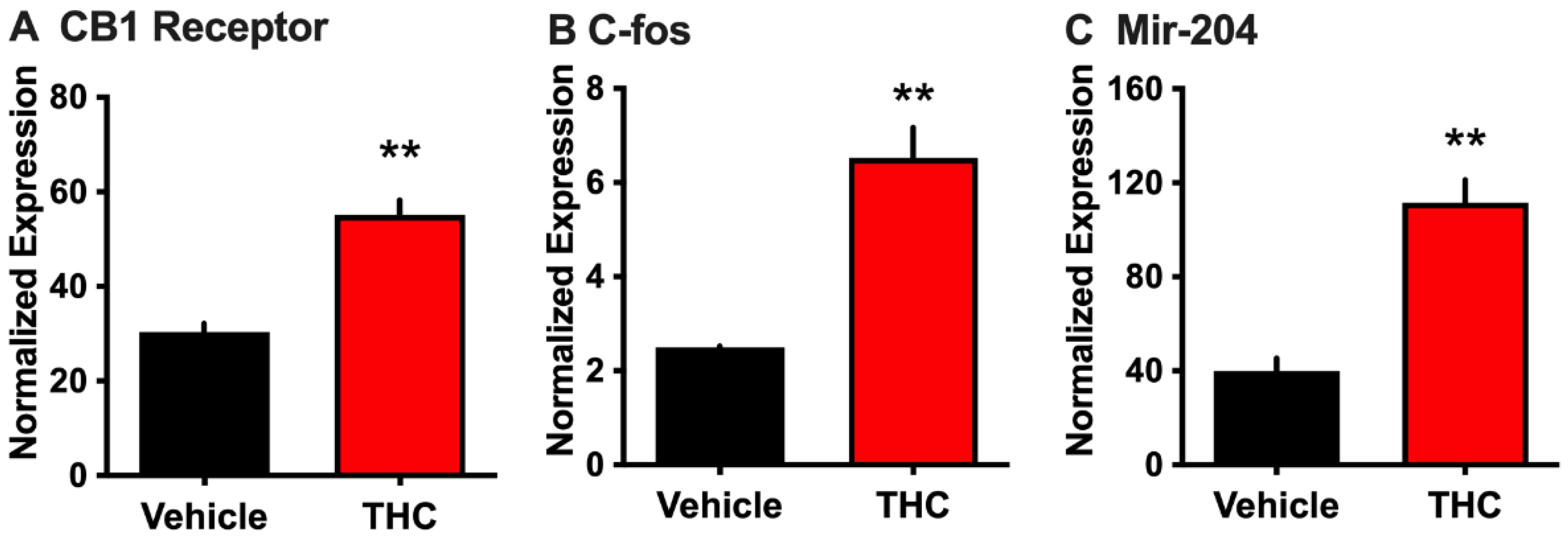
3.1. Effect of THC on Choroid Plexus Epithelial Cells in Primary Culture
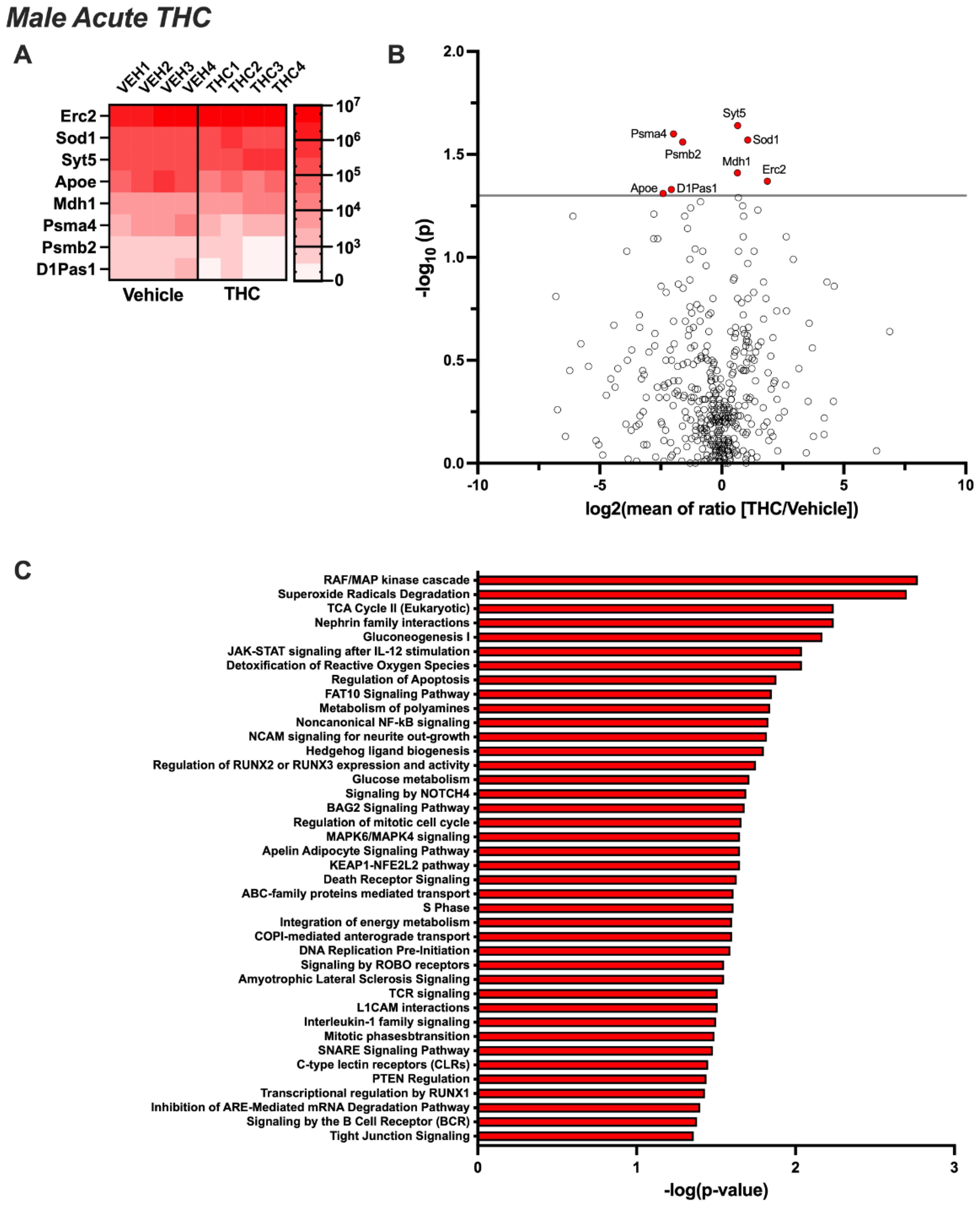
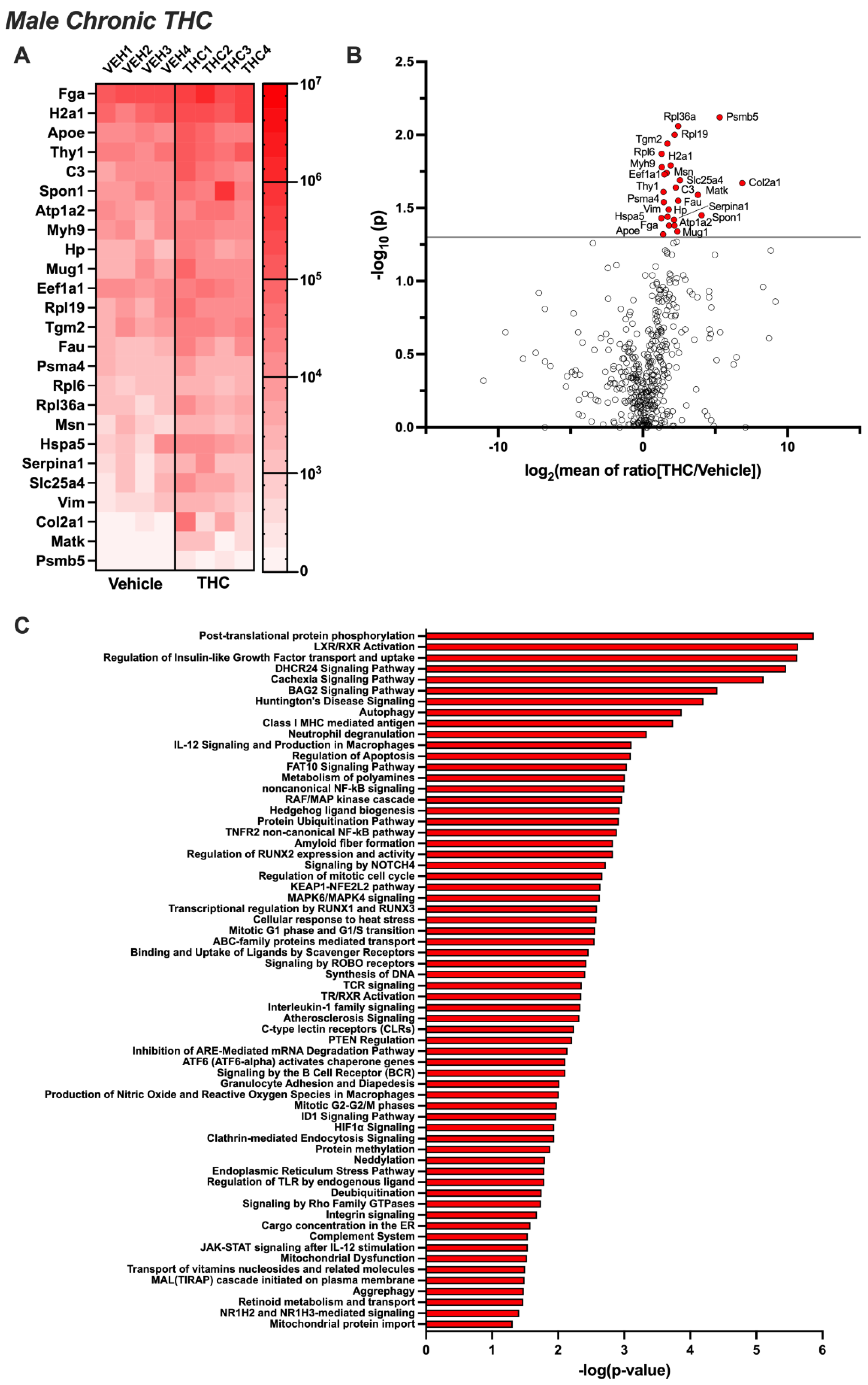
3.2. Differentially Expressed EV Proteins in the Male CSF with Acute or Chronic THC Exposure
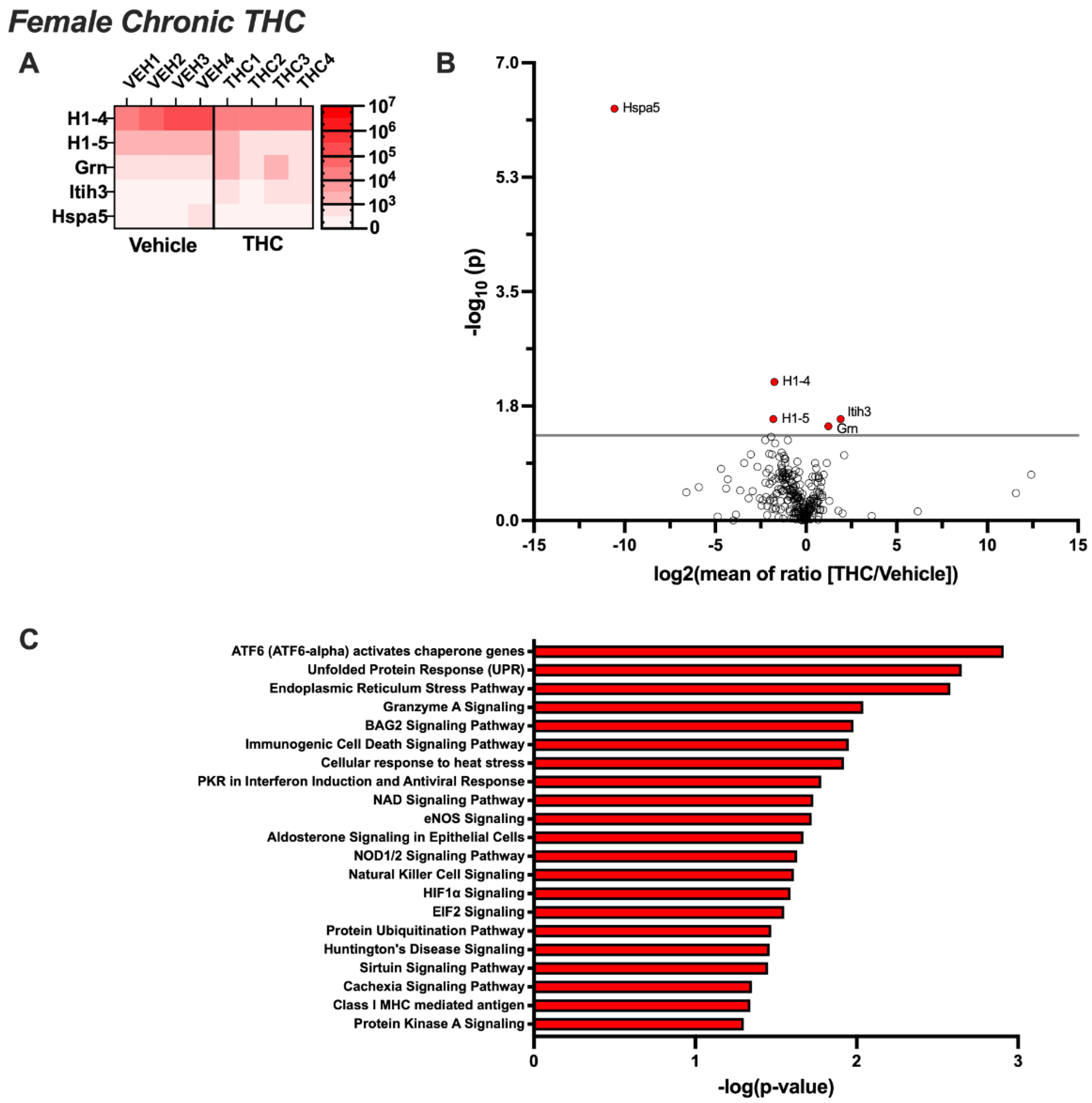
3.3. Differentially Expressed EV Proteins in the Female CSF with Acute or Chronic THC Exposure
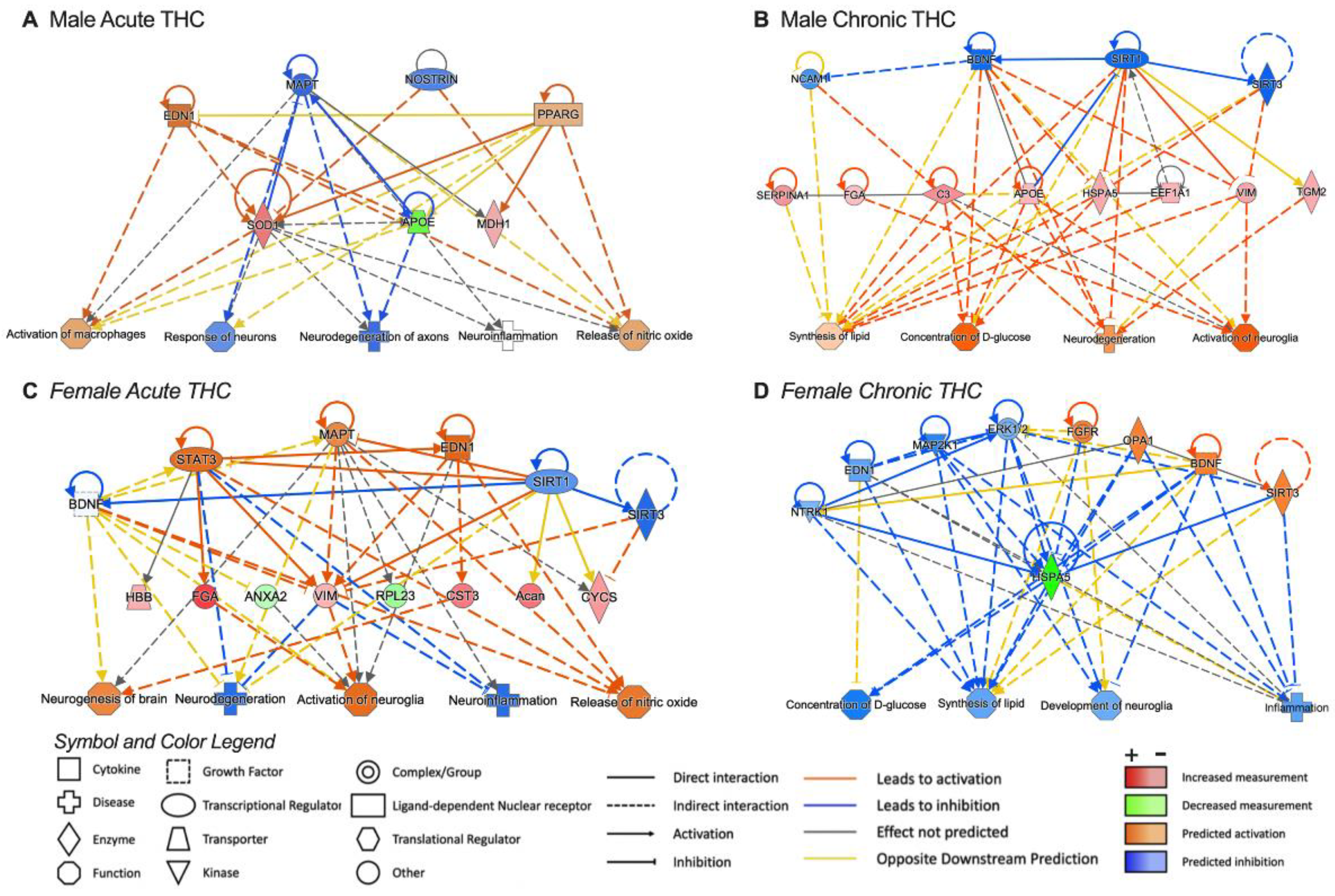
3.4. Pathway Analysis for Significant Targets
4. Discussion
4.1. Relevance of CSF-Derived Extracellular Vesicles
4.2. Interactions Based on THC Exposure Duration and/or Sex
4.3. Putative Significance of Differentially Regulated Proteins
4.3.1. Immune Function and Vesicular Trafficking
4.3.2. Epigenetic Regulators
4.3.3. Experimental Considerations and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gowing, L.R.; Ali, R.L.; Allsop, S.; Marsden, J.; Turf, E.E.; West, R.; Witton, J. Global statistics on addictive behaviours: 2014 status report. Addiction 2015, 110, 904–919. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Srisuma, S.; Bronstein, A.C.; Hoyte, C.O. Characterization of edible marijuana product exposures reported to United States poison centers. Clin. Toxicol. 2016, 54, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.T.; Hill, M.N.; Lee, F.S. Developmental regulation of fear learning and anxiety behavior by endocannabinoids. Genes Brain Behav. 2016, 15, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Renard, J.; Krebs, M.O.; Le Pen, G.; Jay, T.M. Long-term consequences of adolescent cannabinoid exposure in adult psychopathology. Front. Neurosci. 2014, 8, 361. [Google Scholar] [CrossRef]
- Grotenhermen, F. Pharmacokinetics and pharmacodynamics of cannabinoids. Clin. Pharmacokinet. 2003, 42, 327–360. [Google Scholar] [CrossRef] [PubMed]
- Vozella, V.; Zibardi, C.; Ahmed, F.; Piomelli, D. Fast and Sensitive Quantification of Delta(9)-Tetrahydrocannabinol and Its Main Oxidative Metabolites by Liquid Chromatography/Tandem Mass Spectrometry. Cannabis Cannabinoid Res. 2019, 4, 110–123. [Google Scholar] [CrossRef]
- Nguyen, J.D.; Aarde, S.M.; Vandewater, S.A.; Grant, Y.; Stouffer, D.G.; Parsons, L.H.; Cole, M.; Taffe, M.A. Inhaled delivery of Delta(9)-tetrahydrocannabinol (THC) to rats by e-cigarette vapor technology. Neuropharmacology 2016, 109, 112–120. [Google Scholar] [CrossRef]
- Torrens, A.; Ruiz, C.M.; Martinez, M.X.; Tagne, A.M.; Roy, P.; Grimes, D.; Ahmed, F.; Lallai, V.; Inshishian, V.; Bautista, M.; et al. Nasal accumulation and metabolism of Delta(9)-tetrahydrocannabinol following aerosol (‘vaping’) administration in an adolescent rat model. Pharmacol. Res. 2023, 187, 106600. [Google Scholar] [CrossRef]
- Brown, P.D.; Davies, S.L.; Speake, T.; Millar, I.D. Molecular mechanisms of cerebrospinal fluid production. Neuroscience 2004, 129, 957–970. [Google Scholar] [CrossRef]
- Grapp, M.; Wrede, A.; Schweizer, M.; Huwel, S.; Galla, H.J.; Snaidero, N.; Simons, M.; Buckers, J.; Low, P.S.; Urlaub, H.; et al. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat. Commun. 2013, 4, 2123. [Google Scholar] [CrossRef]
- Li, M.D.; Kane, J.K.; Matta, S.G.; Blaner, W.S.; Sharp, B.M. Nicotine enhances the biosynthesis and secretion of transthyretin from the choroid plexus in rats: Implications for beta-amyloid formation. J. Neurosci. 2000, 20, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- Lun, M.P.; Johnson, M.B.; Broadbelt, K.G.; Watanabe, M.; Kang, Y.J.; Chau, K.F.; Springel, M.W.; Malesz, A.; Sousa, A.M.; Pletikos, M.; et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci. 2015, 35, 4903–4916. [Google Scholar] [CrossRef] [PubMed]
- Sawamoto, K.; Wichterle, H.; Gonzalez-Perez, O.; Cholfin, J.A.; Yamada, M.; Spassky, N.; Murcia, N.S.; Garcia-Verdugo, J.M.; Marin, O.; Rubenstein, J.L.; et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 2006, 311, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Aldred, A.R.; Jaworowski, A.; Nilsson, C.; Achen, M.G.; Segal, M.B. Thyroxine transport from blood to brain via transthyretin synthesis in choroid plexus. Am. J. Physiol. 1990, 258, R338–R345. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Bianco, F.; Pravettoni, E.; Colombo, A.; Schenk, U.; Moller, T.; Matteoli, M.; Verderio, C. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J. Immunol. 2005, 174, 7268–7277. [Google Scholar] [CrossRef]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef]
- Colombo, F.; Bastoni, M.; Nigro, A.; Podini, P.; Finardi, A.; Casella, G.; Ramesh, M.; Farina, C.; Verderio, C.; Furlan, R. Cytokines Stimulate the Release of Microvesicles from Myeloid Cells Independently from the P2X7 Receptor/Acid Sphingomyelinase Pathway. Front. Immunol. 2018, 9, 204. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Nakano, I.; Garnier, D.; Minata, M.; Rak, J. Extracellular vesicles in the biology of brain tumour stem cells--Implications for inter-cellular communication, therapy and biomarker development. Semin. Cell Dev. Biol. 2015, 40, 17–26. [Google Scholar] [CrossRef]
- Pastuzyn, E.D.; Day, C.E.; Kearns, R.B.; Kyrke-Smith, M.; Taibi, A.V.; McCormick, J.; Yoder, N.; Belnap, D.M.; Erlendsson, S.; Morado, D.R.; et al. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 2018, 172, 275–288.e18. [Google Scholar] [CrossRef] [PubMed]
- Potolicchio, I.; Carven, G.J.; Xu, X.; Stipp, C.; Riese, R.J.; Stern, L.J.; Santambrogio, L. Proteomic analysis of microglia-derived exosomes: Metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol. 2005, 175, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Budnik, V.; Ruiz-Canada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Fowler, C.D. NeuroEVs: Characterizing Extracellular Vesicles Generated in the Neural Domain. J. Neurosci. 2019, 39, 9262–9268. [Google Scholar] [CrossRef]
- Lallai, V.; Grimes, N.; Fowler, J.P.; Sequeira, P.A.; Cartagena, P.; Limon, A.; Coutts, M.; Monuki, E.S.; Bunney, W.; Demuro, A.; et al. Nicotine Acts on Cholinergic Signaling Mechanisms to Directly Modulate Choroid Plexus Function. eNeuro 2019, 6, e0051. [Google Scholar] [CrossRef]
- Ashton, J.C.; Appleton, I.; Darlington, C.L.; Smith, P.F. Cannabinoid CB1 receptor protein expression in the rat choroid plexus: A possible involvement of cannabinoids in the regulation of cerebrospinal fluid. Neurosci. Lett. 2004, 364, 40–42. [Google Scholar] [CrossRef]
- Suarez, J.; Romero-Zerbo, S.Y.; Rivera, P.; Bermudez-Silva, F.J.; Perez, J.; De Fonseca, F.R.; Fernandez-Llebrez, P. Endocannabinoid system in the adult rat circumventricular areas: An immunohistochemical study. J. Comp. Neurol. 2010, 518, 3065–3085. [Google Scholar] [CrossRef]
- Hunt, C.A.; Jones, R.T. Tolerance and disposition of tetrahydrocannabinol in man. J. Pharmacol. Exp. Ther. 1980, 215, 35–44. [Google Scholar]
- Lallai, V.; Manca, L.; Sherafat, Y.; Fowler, C.D. Effects of Prenatal Nicotine, THC, or Co-Exposure on Cognitive Behaviors in Adolescent Male and Female Rats. Nicotine Tob. Res. 2022, 24, 1150–1160. [Google Scholar] [CrossRef]
- Lallai, V.; Chen, Y.C.; Roybal, M.M.; Kotha, E.R.; Fowler, J.P.; Staben, A.; Cortez, A.; Fowler, C.D. Nicotine e-cigarette vapor inhalation and self-administration in a rodent model: Sex- and nicotine delivery-specific effects on metabolism and behavior. Addict. Biol. 2021, 26, e13024. [Google Scholar] [CrossRef] [PubMed]
- Vidyadhara, D.J.; Somayaji, M.; Wade, N.; Yucel, B.; Zhao, H.; Shashaank, N.; Ribaudo, J.; Gupta, J.; Lam, T.T.; Sames, D.; et al. Dopamine transporter and synaptic vesicle sorting defects underlie auxilin-associated Parkinson’s disease. Cell Rep. 2023, 42, 112231. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Accessed online at: Proteinatlas.org. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]
- Garcia-Contreras, M.; Thakor, A.S. Extracellular vesicles in Alzheimer’s disease: From pathology to therapeutic approaches. Neural Regen. Res. 2023, 18, 18–22. [Google Scholar] [CrossRef]
- Herman, S.; Djaldetti, R.; Mollenhauer, B.; Offen, D. CSF-derived extracellular vesicles from patients with Parkinson’s disease induce symptoms and pathology. Brain 2023, 146, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Afonso, G.J.M.; Cavaleiro, C.; Valero, J.; Mota, S.I.; Ferreiro, E. Recent Advances in Extracellular Vesicles in Amyotrophic Lateral Sclerosis and Emergent Perspectives. Cells 2023, 12, 1763. [Google Scholar] [CrossRef]
- Rather, H.A.; Mishra, S.; Su, Y.; Kumar, A.; Singh, S.; Misra, B.B.; Lee, J.; Furdui, C.M.; Hamilton, L.R.; Gould, R.W.; et al. Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation. Biomolecules 2022, 12, 510. [Google Scholar] [CrossRef]
- Rahman, M.A.; Patters, B.J.; Kodidela, S.; Kumar, S. Extracellular Vesicles: Intercellular Mediators in Alcohol-Induced Pathologies. J. Neuroimmune Pharmacol. 2020, 15, 409–421. [Google Scholar] [CrossRef]
- Chen, F.; Xu, Y.; Shi, K.; Zhang, Z.; Xie, Z.; Wu, H.; Ma, Y.; Zhou, Y.; Chen, C.; Yang, J.; et al. Multi-omics study reveals associations among neurotransmitter, extracellular vesicle-derived microRNA and psychiatric comorbidities during heroin and methamphetamine withdrawal. Biomed. Pharmacother. 2022, 155, 113685. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, H.; Peng, Q.; Xie, Z.; Chen, F.; Ma, Y.; Zhang, Y.; Zhou, Y.; Yang, J.; Chen, C.; et al. Integration of Molecular Inflammatory Interactome Analyses Reveals Dynamics of Circulating Cytokines and Extracellular Vesicle Long Non-Coding RNAs and mRNAs in Heroin Addicts During Acute and Protracted Withdrawal. Front. Immunol. 2021, 12, 730300. [Google Scholar] [CrossRef] [PubMed]
- Sandau, U.S.; Duggan, E.; Shi, X.; Smith, S.J.; Huckans, M.; Schutzer, W.E.; Loftis, J.M.; Janowsky, A.; Nolan, J.P.; Saugstad, J.A. Methamphetamine use alters human plasma extracellular vesicles and their microRNA cargo: An exploratory study. J. Extracell. Vesicles 2020, 10, e12028. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.; Lam, T.T.; Garcia-Milian, R.; Cyril D’Souza, D.; Nairn, A.C.; Elgert, K.; Eitan, E.; Ranganathan, M. Peripheral signature of altered synaptic integrity in young onset cannabis use disorder: A proteomic study of circulating extracellular vesicles. World J. Biol. Psychiatry 2023, 24, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Yamashita, T.; Shi, X.; Feng, T.; Yu, H.; Hu, X.; Hu, X.; Bian, Y.; Sun, H.; Tadokoro, K.; et al. Accelerated accumulation of fibrinogen peptide chains with Abeta deposition in Alzheimer’s disease (AD) mice and human AD brains. Brain Res. 2021, 1767, 147569. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Arboledas, A.; Acharya, M.M.; Tenner, A.J. The Role of Complement in Synaptic Pruning and Neurodegeneration. Immunotargets Ther. 2021, 10, 373–386. [Google Scholar] [CrossRef]
- Nikitidou, E.; Khoonsari, P.E.; Shevchenko, G.; Ingelsson, M.; Kultima, K.; Erlandsson, A. Increased Release of Apolipoprotein E in Extracellular Vesicles Following Amyloid-beta Protofibril Exposure of Neuroglial Co-Cultures. J. Alzheimer’s Dis. 2017, 60, 305–321. [Google Scholar] [CrossRef]
- Jiang, S.; Li, X.; Li, Y.; Chang, Z.; Yuan, M.; Zhang, Y.; Zhu, H.; Xiu, Y.; Cong, H.; Yin, L.; et al. APOE from patient-derived astrocytic extracellular vesicles alleviates neuromyelitis optica spectrum disorder in a mouse model. Sci. Transl. Med. 2024, 16, eadg5116. [Google Scholar] [CrossRef]
- Muraoka, S.; Jedrychowski, M.P.; Iwahara, N.; Abdullah, M.; Onos, K.D.; Keezer, K.J.; Hu, J.; Ikezu, S.; Howell, G.R.; Gygi, S.P.; et al. Enrichment of Neurodegenerative Microglia Signature in Brain-Derived Extracellular Vesicles Isolated from Alzheimer’s Disease Mouse Models. J. Proteome Res. 2021, 20, 1733–1743. [Google Scholar] [CrossRef]
- Mahley, R.W.; Huang, Y. Apolipoprotein e sets the stage: Response to injury triggers neuropathology. Neuron 2012, 76, 871–885. [Google Scholar] [CrossRef]
- Matthaei, A.; Joecks, S.; Frauenstein, A.; Bruening, J.; Bankwitz, D.; Friesland, M.; Gerold, G.; Vieyres, G.; Kaderali, L.; Meissner, F.; et al. Landscape of protein-protein interactions during hepatitis C virus assembly and release. Microbiol. Spectr. 2024, 12, e0256222. [Google Scholar] [CrossRef]
- Zhou, C.; Liu, Y.; Wei, Q.; Chen, Y.; Yang, S.; Cheng, A.; Zhang, G. HSPA5 Promotes Attachment and Internalization of Porcine Epidemic Diarrhea Virus through Interaction with the Spike Protein and the Endo-/Lysosomal Pathway. J. Virol. 2023, 97, e0054923. [Google Scholar] [CrossRef] [PubMed]
- Sornjai, W.; Promma, P.; Priewkhiew, S.; Ramphan, S.; Jaratsittisin, J.; Jinagool, P.; Wikan, N.; Greenwood, M.; Murphy, D.; Smith, D.R. The interaction of GRP78 and Zika virus E and NS1 proteins occurs in a chaperone-client manner. Sci. Rep. 2024, 14, 10407. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Liang, P.; Puthiyakunnon, S.; Yang, L.; Hong, C. VP1-binding protein glucose-regulated protein 78 as an important mediator for enterovirus 71 infecting human brain microvascular endothelial cells. Indian. J. Med. Microbiol. 2019, 37, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.; Roberts, J.L.; Cash, D.R.; Tavallai, S.; Jean, S.; Fidanza, A.; Cruz-Luna, T.; Siembiba, P.; Cycon, K.A.; Cornelissen, C.N.; et al. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J. Cell Physiol. 2015, 230, 1661–1676. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Galvez, D.; Dangerfield, J.A.; Metzner, C. Extracellular Vesicles and Their Membranes: Exosomes vs. Virus-Related Particles. Membranes 2023, 13, 397. [Google Scholar] [CrossRef]
- Arrindell, J.; Desnues, B. Vimentin: From a cytoskeletal protein to a critical modulator of immune response and a target for infection. Front. Immunol. 2023, 14, 1224352. [Google Scholar] [CrossRef]
- Zhao, S.; Miao, C.; Gao, X.; Li, Z.; Eriksson, J.E.; Jiu, Y. Vimentin cage—A double-edged sword in host anti-infection defense. Curr. Opin. Cell Biol. 2024, 86, 102317. [Google Scholar] [CrossRef]
- Ramos, I.; Stamatakis, K.; Oeste, C.L.; Perez-Sala, D. Vimentin as a Multifaceted Player and Potential Therapeutic Target in Viral Infections. Int. J. Mol. Sci. 2020, 21, 4675. [Google Scholar] [CrossRef]
- Boylan, M.A.; Pincetic, A.; Romano, G.; Tatton, N.; Kenkare-Mitra, S.; Rosenthal, A. Targeting Progranulin as an Immuno-Neurology Therapeutic Approach. Int. J. Mol. Sci. 2023, 24, 15946. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, L.; Bracko, O.; Choi, J.W.; Jia, Y.; Nana, A.L.; Brady, O.A.; Hernandez, J.C.C.; Nishimura, N.; Seeley, W.W.; et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat. Commun. 2017, 8, 15277. [Google Scholar] [CrossRef]
- Saegusa, C.; Fukuda, M.; Mikoshiba, K. Synaptotagmin V is targeted to dense-core vesicles that undergo calcium-dependent exocytosis in PC12 cells. J. Biol. Chem. 2002, 277, 24499–24505. [Google Scholar] [CrossRef] [PubMed]
- Vinet, A.F.; Fukuda, M.; Descoteaux, A. The exocytosis regulator synaptotagmin V controls phagocytosis in macrophages. J. Immunol. 2008, 181, 5289–5295. [Google Scholar] [CrossRef] [PubMed]
- Nishimune, H. Active zones of mammalian neuromuscular junctions: Formation, density, and aging. Ann. N. Y. Acad. Sci. 2012, 1274, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Kiyonaka, S.; Nakajima, H.; Takada, Y.; Hida, Y.; Yoshioka, T.; Hagiwara, A.; Kitajima, I.; Mori, Y.; Ohtsuka, T. Physical and functional interaction of the active zone protein CAST/ERC2 and the beta-subunit of the voltage-dependent Ca(2+) channel. J. Biochem. 2012, 152, 149–159. [Google Scholar] [CrossRef]
- Farrelly, L.A.; Thompson, R.E.; Zhao, S.; Lepack, A.E.; Lyu, Y.; Bhanu, N.V.; Zhang, B.; Loh, Y.E.; Ramakrishnan, A.; Vadodaria, K.C.; et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 2019, 567, 535–539. [Google Scholar] [CrossRef]
- Jearjaroen, P.; Thangwong, P.; Tocharus, C.; Chaichompoo, W.; Suksamrarn, A.; Tocharus, J. Hexahydrocurcumin attenuated demyelination and improved cognitive impairment in chronic cerebral hypoperfusion rats. Inflammopharmacology 2024, 32, 1531–1544. [Google Scholar] [CrossRef]
- Rossin, F.; Ciccosanti, F.; D’Eletto, M.; Occhigrossi, L.; Fimia, G.M.; Piacentini, M. Type 2 transglutaminase in the nucleus: The new epigenetic face of a cytoplasmic enzyme. Cell. Mol. Life Sci. 2023, 80, 52. [Google Scholar] [CrossRef]
- Lepack, A.E.; Werner, C.T.; Stewart, A.F.; Fulton, S.L.; Zhong, P.; Farrelly, L.A.; Smith, A.C.W.; Ramakrishnan, A.; Lyu, Y.; Bastle, R.M.; et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 2020, 368, 197–201. [Google Scholar] [CrossRef]
- Holmberg Olausson, K.; Elsir, T.; Moazemi Goudarzi, K.; Nister, M.; Lindstrom, M.S. NPM1 histone chaperone is upregulated in glioblastoma to promote cell survival and maintain nucleolar shape. Sci. Rep. 2015, 5, 16495. [Google Scholar] [CrossRef]
- Sang, B.; Sun, J.; Yang, D.; Xu, Z.; Wei, Y. Ras-AKT signaling represses the phosphorylation of histone H1.5 at threonine 10 via GSK3 to promote the progression of glioma. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2882–2890. [Google Scholar] [CrossRef]
- Agliardi, C.; Clerici, M. Blood extracellular vesicles (EVs) of central nervous system origin: A window into the brain. Neural Regen. Res. 2020, 15, 55–56. [Google Scholar] [CrossRef] [PubMed]
- Balusu, S.; Van Wonterghem, E.; De Rycke, R.; Raemdonck, K.; Stremersch, S.; Gevaert, K.; Brkic, M.; Demeestere, D.; Vanhooren, V.; Hendrix, A.; et al. Identification of a novel mechanism of blood-brain communication during peripheral inflammation via choroid plexus-derived extracellular vesicles. EMBO Mol. Med. 2016, 8, 1162–1183. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.G.; Gray, E.; Heman-Ackah, S.M.; Mager, I.; Talbot, K.; Andaloussi, S.E.; Wood, M.J.; Turner, M.R. Extracellular vesicles in neurodegenerative disease—Pathogenesis to biomarkers. Nat. Rev. Neurol. 2016, 12, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Schnatz, A.; Muller, C.; Brahmer, A.; Kramer-Albers, E.M. Extracellular Vesicles in neural cell interaction and CNS homeostasis. FASEB Bioadv. 2021, 3, 577–592. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lallai, V.; Lam, T.T.; Garcia-Milian, R.; Chen, Y.-C.; Fowler, J.P.; Manca, L.; Piomelli, D.; Williams, K.; Nairn, A.C.; Fowler, C.D. Proteomic Profile of Circulating Extracellular Vesicles in the Brain after Δ9-Tetrahydrocannabinol Inhalation. Biomolecules 2024, 14, 1143. https://doi.org/10.3390/biom14091143
Lallai V, Lam TT, Garcia-Milian R, Chen Y-C, Fowler JP, Manca L, Piomelli D, Williams K, Nairn AC, Fowler CD. Proteomic Profile of Circulating Extracellular Vesicles in the Brain after Δ9-Tetrahydrocannabinol Inhalation. Biomolecules. 2024; 14(9):1143. https://doi.org/10.3390/biom14091143
Chicago/Turabian StyleLallai, Valeria, TuKiet T. Lam, Rolando Garcia-Milian, Yen-Chu Chen, James P. Fowler, Letizia Manca, Daniele Piomelli, Kenneth Williams, Angus C. Nairn, and Christie D. Fowler. 2024. "Proteomic Profile of Circulating Extracellular Vesicles in the Brain after Δ9-Tetrahydrocannabinol Inhalation" Biomolecules 14, no. 9: 1143. https://doi.org/10.3390/biom14091143






