SUMOylation in Drosophila Development
Abstract
:1. The SUMO Conjugation Pathway in Drosophila
2. Roles for SUMO in Chromosome/Chromatin Function
2.1. Roles in the Meiotic and Mitotic Chromosome Cycles
2.2. Roles in Insulator Function
2.3. Connections to Heterochromatic Silencing
2.4. Links to Polycomb Group Function

3. A Role for SUMO in Eggshell Patterning
4. Embryonic Patterning
4.1. Anteroposterior Patterning
4.2. Dorsoventral Patterning

5. Wing Morphogenesis
5.1. Regulation of Vestigial Function by SUMO
5.2. Regulation of Sal and Salr by SUMO
5.3. Regulation of Homeodomain interacting protein kinase (Hipk) by SUMO

6. Metamorphosis
7. Neurogenesis
8. Innate Immunity
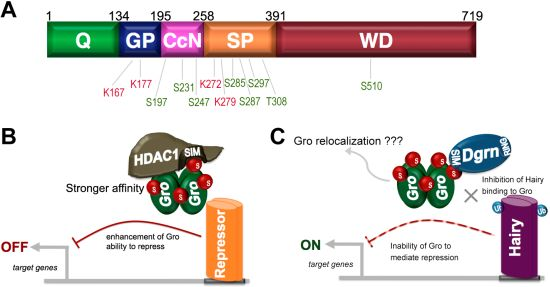
9. SUMO as a Regulator of Groucho Function

10. A Potential Role for p53 SUMOylation in Apoptosis
11. Conclusions
References
- Kumar, D.; Misra, J.R.; Kumar, A.; Chugh, J.; Sharma, S.; Hosur, R.V. NMR-derived solution structure of SUMO from Drosophila melanogaster (dSmt3). Proteins 2009, 75, 1046–1050. [Google Scholar]
- Bohren, K.M.; Nadkarni, V.; Song, J.H.; Gabbay, K.H.; Owerbach, D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J. Biol. Chem. 2004, 279, 27233–27238. [Google Scholar]
- Saitoh, H.; Hinchey, J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000, 275, 6252–6258. [Google Scholar] [CrossRef]
- Huang, H.W.; Tsoi, S.C.; Sun, Y.H.; Li, S.S. Identification and characterization of the SMT3 cDNA and gene encoding ubiquitin-like protein from Drosophila melanogaster. Biochem. Mol. Biol. Int. 1998, 46, 775–785. [Google Scholar]
- Smith, M.; Bhaskar, V.; Fernandez, J.; Courey, A.J. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugate. J. Biol. Chem. 2004, 279, 43805–43814. [Google Scholar]
- Long, X.; Griffith, L.C. Identification and characterization of a SUMO-1 conjugation system that modifies neuronal calcium/calmodulin-dependent protein kinase II in Drosophila melanogaster. J. Biol. Chem. 2000, 275, 40765–40776. [Google Scholar]
- Rodriguez, M.S.; Dargemont, C.; Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001, 276, 12654–12659. [Google Scholar]
- Ulrich, H.D. The fast-growing business of SUMO chains. Mol. Cell 2008, 32, 301–305. [Google Scholar] [CrossRef]
- Stielow, B.; Sapetschnig, A.; Kruger, I.; Kunert, N.; Brehm, A.; Boutros, M.; Suske, G. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 2008, 29, 742–754. [Google Scholar] [CrossRef]
- Betz, A.; Lampen, N.; Martinek, S.; Young, M.W.; Darnell, J.E., Jr. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 2001, 98, 9563–9568. [Google Scholar]
- Apionishev, S.; Malhotra, D.; Raghavachari, S.; Tanda, S.; Rasooly, R.S. The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells 2001, 6, 215–224. [Google Scholar] [CrossRef]
- Nie, M.; Xie, Y.; Loo, J.A.; Courey, A.J. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PloS One 2009, 4, e5905. [Google Scholar]
- Capelson, M.; Corces, V.G. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. EMBO J. 2006, 25, 1906–1914. [Google Scholar] [CrossRef]
- Capelson, M.; Corces, V.G. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 2005, 20, 105–116. [Google Scholar] [CrossRef]
- Matsui, M.; Sharma, K.C.; Cooke, C.; Wakimoto, B.T.; Rasool, M.; Hayworth, M.; Hylton, C.A.; Tomkiel, J.E. Nuclear structure and chromosome segregation in Drosophila male meiosis depend on the ubiquitin ligase dTopors. Genetics 2011, 189, 779–793. [Google Scholar]
- Golovnin, A.; Volkov, I.; Georgiev, P. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J. Cell Sci. 2012, 125, 2064–2074. [Google Scholar]
- Schotta, G.; Ebert, A.; Krauss, V.; Fischer, A.; Hoffmann, J.; Rea, S.; Jenuwein, T.; Dorn, R.; Reuter, G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002, 21, 1121–1131. [Google Scholar]
- Hari, K.L.; Cook, K.R.; Karpen, G.H. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001, 15, 1334–1348. [Google Scholar]
- Reo, E.; Seum, C.; Spierer, P.; Bontron, S. Sumoylation of Drosophila SU(VAR)3-7 is required for its heterochromatic function. Nucleic Acids Res. 2010, 38, 4254–4262. [Google Scholar]
- Simon, J.A.; Kingston, R.E. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar]
- Smith, M.; Mallin, D.R.; Simon, J.A.; Courey, A.J. Small ubiquitin-like modifier (SUMO) conjugation impedes transcriptional silencing by the polycomb group repressor Sex Comb on Midleg. J. Biol. Chem. 2011, 286, 11391–11400. [Google Scholar]
- Lewis, E.B. The bithorax complex: The first fifty years. Int. J. Dev. Biol. 1998, 42, 403–415. [Google Scholar]
- Kagey, M.H.; Melhuish, T.A.; Wotton, D. The polycomb protein Pc2 is a SUMO E3. Cell 2003, 113, 127–137. [Google Scholar]
- Schnorr, J.D.; Holdcraft, R.; Chevalier, B.; Berg, C.A. Ras1 interacts with multiple new signaling and cytoskeletal loci in Drosophila eggshell patterning and morphogenesis. Genetics 2001, 159, 609–622. [Google Scholar]
- Schweitzer, R.; Shilo, B.Z. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997, 13, 191–196. [Google Scholar] [CrossRef]
- Lehembre, F.; Badenhorst, P.; Muller, S.; Travers, A.; Schweisguth, F.; Dejean, A. Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol. Cell Biol. 2000, 20, 1072–1082. [Google Scholar]
- Hashiyama, K.; Shigenobu, S.; Kobayashi, S. Expression of genes involved in sumoylation in the Drosophila germline. Gene Expr. Patterns 2009, 9, 50–53. [Google Scholar] [CrossRef]
- Epps, J.L.; Tanda, S. The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 1998, 8, 1277–1280. [Google Scholar] [CrossRef]
- Liu, J.; Ma, J. Drosophila Bicoid is a substrate of sumoylation and its activator function is subject to inhibition by this post-translational modification. FEBS Lett. 2012, 586, 1719–1723. [Google Scholar] [CrossRef]
- Bhaskar, V.; Smith, M.; Courey, A.J. Conjugation of Smt3 to dorsal may potentiate the Drosophila immune response. Mol. Cell Biol. 2002, 22, 492–504. [Google Scholar] [CrossRef]
- Bhaskar, V.; Valentine, S.A.; Courey, A.J. A functional interaction between dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem. 2000, 275, 4033–4040. [Google Scholar]
- Miles, W.O.; Jaffray, E.; Campbell, S.G.; Takeda, S.; Bayston, L.J.; Basu, S.P.; Li, M.; Raftery, L.A.; Ashe, M.P.; Hay, R.T.; et al. Medea SUMOylation restricts the signaling range of the Dpp morphogen in the Drosophila embryo. Genes Dev. 2008, 22, 2578–2590. [Google Scholar] [CrossRef]
- Affolter, M.; Marty, T.; Vigano, M.A.; Jazwinska, A. Nuclear interpretation of Dpp signaling in Drosophila. EMBO J. 2001, 20, 3298–3305. [Google Scholar] [CrossRef]
- Gao, S.; Steffen, J.; Laughon, A. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 2005, 280, 36158–36164. [Google Scholar]
- Long, J.; Wang, G.; He, D.; Liu, F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 2004, 379, 23–29. [Google Scholar] [CrossRef]
- Takanaka, Y.; Courey, A.J. SUMO enhances vestigial function during wing morphogenesis. Mech. Dev. 2005, 122, 1130–1137. [Google Scholar] [CrossRef]
- De Celis, J.F.; Barrio, R. Regulation and function of Spalt proteins during animal development. Int. J. Dev. Biol. 2009, 53, 1385–1398. [Google Scholar] [CrossRef]
- Netzer, C.; Bohlander, S.K.; Rieger, L.; Muller, S.; Kohlhase, J. Interaction of the developmental regulator SALL1 with UBE2I and SUMO-1. Biochem. Biophys. Res. Commun. 2002, 296, 870–876. [Google Scholar] [CrossRef]
- Sanchez, J.; Talamillo, A.; Lopitz-Otsoa, F.; Perez, C.; Hjerpe, R.; Sutherland, J.D.; Herboso, L.; Rodriguez, M.S.; Barrio, R. Sumoylation modulates the activity of Spalt-like proteins during wing development in Drosophila. J. Biol. Chem. 2010, 285, 25841–25849. [Google Scholar]
- Sanchez, J.; Talamillo, A.; Gonzalez, M.; Sanchez-Pulido, L.; Jimenez, S.; Pirone, L.; Sutherland, J.D.; Barrio, R. Drosophila Sal and Salr are transcriptional repressors. Biochem. J. 2011, 438, 437–445. [Google Scholar]
- Hofmann, T.G.; Stollberg, N.; Schmitz, M.L.; Will, H. HIPK2 regulates transforming growth factor-beta-induced c-Jun NH(2)-terminal kinase activation and apoptosis in human hepatoma cells. Cancer Res. 2003, 63, 8271–8277. [Google Scholar]
- Lan, H.C.; Li, H.J.; Lin, G.; Lai, P.Y.; Chung, B.C. Cyclic AMP stimulates SF-1-dependent CYP11A1 expression through homeodomain-interacting protein kinase 3-mediated Jun N-terminal kinase and c-Jun phosphorylation. Mol. Cell Biol. 2007, 27, 2027–2036. [Google Scholar] [CrossRef]
- Huang, H.; Du, G.; Chen, H.; Liang, X.; Li, C.; Zhu, N.; Xue, L.; Ma, J.; Jiao, R. Drosophila Smt3 negatively regulates JNK signaling through sequestering Hipk in the nucleus. Development 2011, 138, 2477–2485. [Google Scholar]
- Chiu, H.; Ring, B.C.; Sorrentino, R.P.; Kalamarz, M.; Garza, D.; Govind, S. dUbc9 negatively regulates the Toll-NF-kappa B pathways in larval hematopoiesis and drosomycin activation in Drosophila. Dev. Biol. 2005, 288, 60–72. [Google Scholar] [CrossRef]
- Talamillo, A.; Sanchez, J.; Cantera, R.; Perez, C.; Martin, D.; Caminero, E.; Barrio, R. Smt3 is required for Drosophila melanogaster metamorphosis. Development 2008, 135, 1659–1668. [Google Scholar] [CrossRef]
- Warren, J.T.; Petryk, A.; Marques, G.; Parvy, J.P.; Shinoda, T.; Itoyama, K.; Kobayashi, J.; Jarcho, M.; Li, Y.; O’Connor, M.B.; et al. Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: A P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 2004, 34, 991–1010. [Google Scholar] [CrossRef]
- Guo, M.; Jan, L.Y.; Jan, Y.N. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron 1996, 17, 27–41. [Google Scholar] [CrossRef]
- Savare, J.; Bonneaud, N.; Girard, F. SUMO represses transcriptional activity of the Drosophila SoxNeuro and human Sox3 central nervous system-specific transcription factors. Mol. Biol. Cell 2005, 16, 2660–2669. [Google Scholar] [CrossRef]
- Powell, L.M.; Chen, A.; Huang, Y.C.; Wang, P.Y.; Kemp, S.E.; Jarman, A.P. The SUMO pathway promotes bHLH proneural factor activity via a direct effect on the Zn finger protein, Senseless. Mol. Cell Biol. 2012. [Google Scholar] [CrossRef]
- Huang, L.; Ohsako, S.; Tanda, S. The lesswright mutation activates Rel-related proteins, leading to overproduction of larval hemocytes in Drosophila melanogaster. Dev. Biol. 2005, 280, 407–420. [Google Scholar] [CrossRef]
- Paddibhatla, I.; Lee, M.J.; Kalamarz, M.E.; Ferrarese, R.; Govind, S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010, 6, e1001234. [Google Scholar] [CrossRef]
- Desterro, J.M.; Rodriguez, M.S.; Hay, R.T. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 1998, 2, 233–239. [Google Scholar] [CrossRef]
- Turki-Judeh, W.; Courey, A.J. Groucho a corepressor with instructive roles in development. Curr. Top. Dev. Biol. 2012, 98, 65–96. [Google Scholar] [CrossRef]
- Stifani, S.; Blaumueller, C.M.; Redhead, N.J.; Hill, R.E.; Artavanis-Tsakonas, S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 1992, 2, 343. [Google Scholar]
- Turki-Judeh, W.; Courey, A.J. The Unconserved Groucho Central Region is essential for Viability and Modulates Target Gene Specificity. PloS One 2012, 7, e30610. [Google Scholar]
- Ahn, J.W.; Lee, Y.A.; Ahn, J.H.; Choi, C.Y. Covalent conjugation of Groucho with SUMO-1 modulates its corepressor activity. Biochem. Biophys. Res. Commun. 2009, 379, 160–165. [Google Scholar] [CrossRef]
- Abed, M.; Barry, K.C.; Kenyagin, D.; Koltun, B.; Phippen, T.M.; Delrow, J.J.; Parkhurst, S.M.; Orian, A. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011, 30, 1289–1301. [Google Scholar] [CrossRef]
- Abed, M.; Bitman-Lotan, E.; Orian, A. A fly view of a SUMO-targeted ubiquitin ligase. Fly (Austin) 2011, 5, 340–344. [Google Scholar]
- Wu, S.Y.; Chiang, C.M. p53 sumoylation: Mechanistic insights from reconstitution studies. Epigenetics Off. J. DNA Methylation Soc. 2009, 4, 445–451. [Google Scholar]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef]
- Melchior, F.; Hengst, L. SUMO-1 and p53. Cell Cycle 2002, 1, 245–249. [Google Scholar]
- Muller, S.; Ledl, A.; Schmidt, D. SUMO: A regulator of gene expression and genome integrity. Oncogene 2004, 23, 1998–2008. [Google Scholar] [CrossRef]
- Mauri, F.; McNamee, L.M.; Lunardi, A.; Chiacchiera, F.; Del Sal, G.; Brodsky, M.H.; Collavin, L. Modification of Drosophila p53 by SUMO modulates its transactivation and pro-apoptotic functions. J. Biol. Chem. 2008, 283, 20848–20856. [Google Scholar]
- Formstecher, E.; Aresta, S.; Collura, V.; Hamburger, A.; Meil, A.; Trehin, A.; Reverdy, C.; Betin, V.; Maire, S.; Brun, C.; et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005, 15, 376–384. [Google Scholar] [CrossRef]
- Stanyon, C.A.; Liu, G.; Mangiola, B.A.; Patel, N.; Giot, L.; Kuang, B.; Zhang, H.; Zhong, J.; Finley, R.L., Jr. A drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004, 5, R96. [Google Scholar] [CrossRef]
- Pardi, N.; Vamos, E.; Ujfaludi, Z.; Komonyi, O.; Bodai, L.; Boros, I.M. In vivo effects of abolishing the single canonical sumoylation site in the C-terminal region of Drosophila p53. Acta Biol. Hung. 2011, 62, 397–412. [Google Scholar] [CrossRef]
- Muller, S.; Berger, M.; Lehembre, F.; Seeler, J.S.; Haupt, Y.; Dejean, A. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 2000, 275, 13321–13329. [Google Scholar]
- Feng, L.; Lin, T.; Uranishi, H.; Gu, W.; Xu, Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell Biol. 2005, 25, 5389–5395. [Google Scholar]
- Lomeli, H.; Vazquez, M. Emerging roles of the SUMO pathway in development. Cell Mol. Life Sci. 2011, 68, 4045–4064. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Smith, M.; Turki-Judeh, W.; Courey, A.J. SUMOylation in Drosophila Development. Biomolecules 2012, 2, 331-349. https://doi.org/10.3390/biom2030331
Smith M, Turki-Judeh W, Courey AJ. SUMOylation in Drosophila Development. Biomolecules. 2012; 2(3):331-349. https://doi.org/10.3390/biom2030331
Chicago/Turabian StyleSmith, Matthew, Wiam Turki-Judeh, and Albert J. Courey. 2012. "SUMOylation in Drosophila Development" Biomolecules 2, no. 3: 331-349. https://doi.org/10.3390/biom2030331
APA StyleSmith, M., Turki-Judeh, W., & Courey, A. J. (2012). SUMOylation in Drosophila Development. Biomolecules, 2(3), 331-349. https://doi.org/10.3390/biom2030331