Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures
Abstract
:1. Introduction
2. Results
2.1. Production, Purification and Characterization of EVs

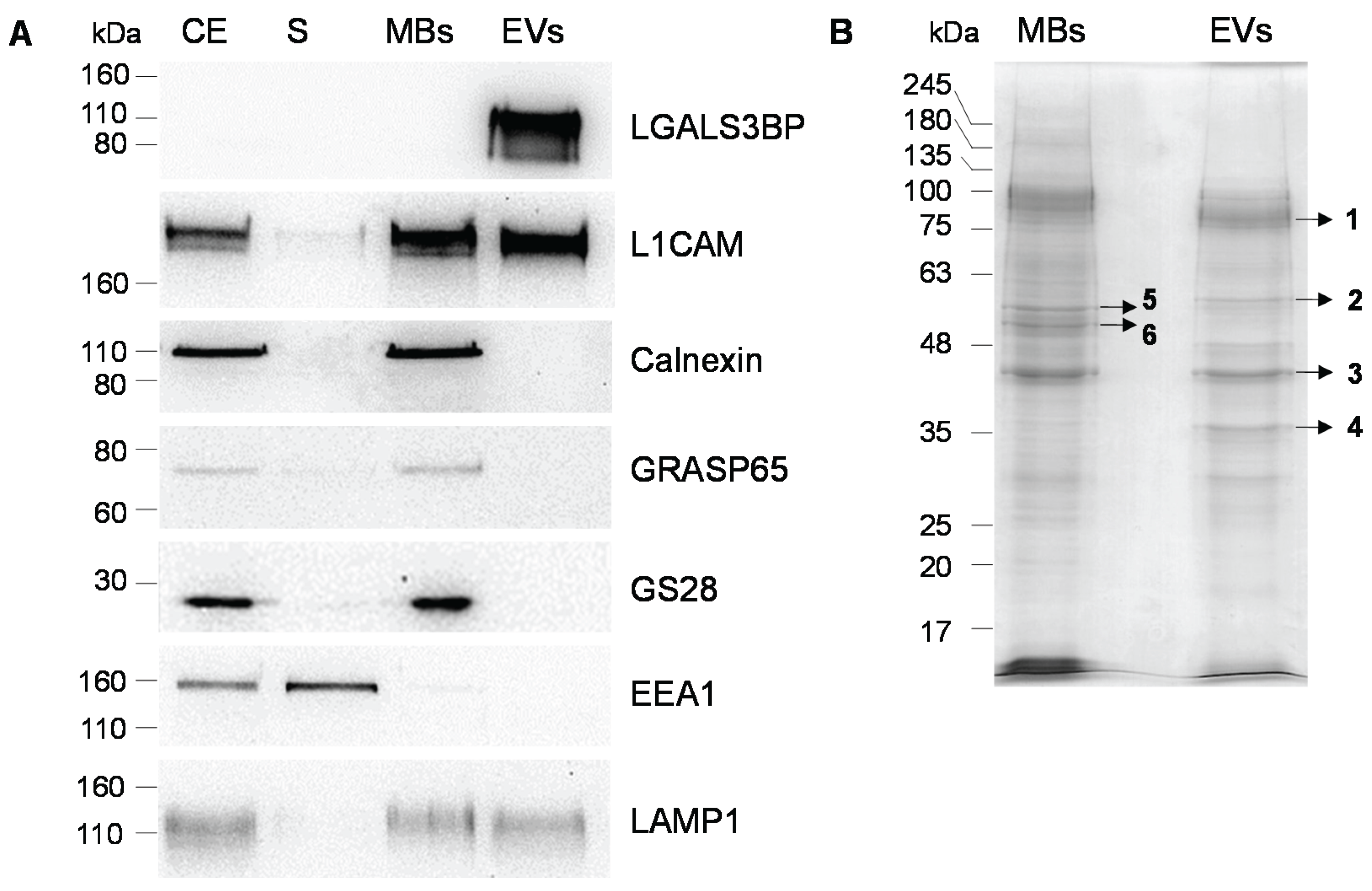
| Gel Band | Protein Name | UniProt Identifier | Gene Name | Nominal Mass (Mr) | Protein Score | Sequence Coverage (%) | Queries Matched | Vesiclepedia |
|---|---|---|---|---|---|---|---|---|
| 1 | Galectin-3-binding protein | LG3BP_HUMAN | LGALS3BP | 65,289 | 460 | 27 | 13 | + |
| Alpha-actinin-4 | ACTN4_HUMAN | ACTN4 | 104,788 | 50 | 10 | 8 | + | |
| 2 | Pyruvate kinase PKM | KPYM_HUMAN | PKM | 57,900 | 47 | 18 | 7 | + |
| 3 | Actin, cytoplasmic 2 | ACTG_HUMAN | ACTG1 | 41,766 | 447 | 50 | 15 | + |
| Actin, alpha cardiac muscle | ACTC_HUMAN | ACTC1 | 41,992 | 166 | 19 | 7 | + | |
| Actin, alpha skeletal muscle | ACTS_HUMAN | ACTA1 | 42,024 | + | ||||
| 4 | Glyceraldehyde-3-phosphate dehydrogenase | G3P_HUMAN | GAPDH | 36,030 | 157 | 52 | 16 | + |
| Ezrin | EZRI_HUMAN | EZR | 69,370 | 69 | 3 | 3 | + | |
| Moesin | MOES_HUMAN | MSN | 67,778 | + | ||||
| Radixin | RADI_HUMAN | RDX | 68,521 | + | ||||
| 5 | Vimentin | VIME_HUMAN | VIM | 53,619 | 415 | 48 | 21 | + |
| 6 | Keratin, type II cytoskeletal 8 | K2C8_HUMAN | KRT8 | 53,671 | 84 | 21 | 12 | + |
| Tubulin alpha-1B chain | TBA1B_HUMAN | TUBA1B | 50,120 | 74 | 22 | 7 | + | |
| Tubulin beta-4A chain | TBB4A_HUMAN | TUBB4A | 49,554 | 54 | 18 | 6 | + | |
| Tubulin beta-4B chain | TBB4B_HUMAN | TUBB4B | 49,799 | + |

2.2. Glycosignatures of EVs and MBs

2.3. Effect of the Glycosylation Inhibitor Kifunensine

3. Discussion
3.1. EVs Glycosignatures and Glycoprotein Sorting Mechanisms
3.2. EVs Glycosignatures and Disease Diagnosis
3.3. EVs Glycosignatures and EVs Uptake by Other Cells
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Cellular Extract, Extracellular Vesicles and Total Cell Membranes
4.3. Immunoblotting and Lectin Blotting Analysis
4.4. Immunoprecipitation and Deglycosylation of LGALS3BP
4.5. Nanoparticle Tracking Analysis (NTA)
4.6. MALDI-TOF/TOF Analysis and Protein Identification
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015. [Google Scholar] [CrossRef] [PubMed]
- Marleau, A.M.; Chen, C.S.; Joyce, J.A.; Tullis, R.H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med. 2012. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Specific tetraspanin functions. J. Cell Biol. 2001, 155, 1103–1107. [Google Scholar] [CrossRef] [PubMed]
- Berditchevski, F. Complexes of tetraspanins with integrins: More than meets the eye. J. Cell Sci. 2001, 114, 4143–4151. [Google Scholar] [PubMed]
- Andreu, Z.; Yanez-Mo, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Perez-Hernandez, D.; Gutierrez-Vazquez, C.; Jorge, I.; Lopez-Martin, S.; Ursa, A.; Sanchez-Madrid, F.; Vazquez, J.; Yanez-Mo, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, L.; Bess, J.W., Jr.; Preston, A.B.; Nagashima, K.; Mahal, L.K. HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat. Chem. Biol. 2009, 5, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Batista, B.S.; Eng, W.S.; Pilobello, K.T.; Hendricks-Munoz, K.D.; Mahal, L.K. Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 2011, 10, 4624–4633. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Grammel, N.; Kandzia, S.; Zeiser, J.; Tranfield, E.M.; Conradt, H.S.; Costa, J. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS ONE 2013, 8, e78631. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta 2012, 1826, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Eng, W.S.; Colquhoun, D.R.; Dinglasan, R.R.; Graham, D.R.; Mahal, L.K. Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J. Biol. Chem. 2014, 289, 32526–32537. [Google Scholar] [CrossRef] [PubMed]
- Gutwein, P.; Stoeck, A.; Riedle, S.; Gast, D.; Runz, S.; Condon, T.P.; Marme, A.; Phong, M.C.; Linderkamp, O.; Skorokhod, A.; et al. Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clin. Cancer Res. 2005, 11, 2492–2501. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Brakebusch, C.; Engel, J.; Timpl, R. Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. EMBO J. 1998, 17, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Etzler, M.E. Antibodies and Lectins in Glycan Analysis. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Abbott, K.L.; Lim, J.M.; Wells, L.; Benigno, B.B.; McDonald, J.F.; Pierce, M. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 2010, 10, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Allam, H.; Aoki, K.; Benigno, B.B.; McDonald, J.F.; Mackintosh, S.G.; Tiemeyer, M.; Abbott, K.L. Glycomic analysis of membrane glycoproteins with bisecting glycosylation from ovarian cancer tissues reveals novel structures and functions. J. Proteome Res. 2015, 14, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; Kandzia, S.; Carilho, R.; Altevogt, P.; Conradt, H.S.; Costa, J. N-Glycosylation of total cellular glycoproteins from the human ovarian carcinoma SKOV3 cell line and of recombinantly expressed human erythropoietin. Glycobiology 2011, 21, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Barres, C.; Blanc, L.; Bette-Bobillo, P.; Andre, S.; Mamoun, R.; Gabius, H.J.; Vidal, M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Block, A.S.; Saraswati, S.; Lichti, C.F.; Mahadevan, M.; Diekman, A.B. Co-purification of Mac-2 binding protein with galectin-3 and association with prostasomes in human semen. Prostate 2011, 71, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Delacour, D.; Koch, A.; Jacob, R. The role of galectins in protein trafficking. Traffic 2009, 10, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Mo, D.; Costa, S.A.; Ihrke, G.; Youker, R.T.; Pastor-Soler, N.; Hughey, R.P.; Weisz, O.A. Sialylation of N-linked glycans mediates apical delivery of endolyn in MDCK cells via a galectin-9-dependent mechanism. Mol. Biol. Cell 2012, 23, 3636–3646. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Howe, A.G.; Xu, G.; Frohlich, O.; Klein, J.D.; Sands, J.M. Mature N-linked glycans facilitate UT-A1 urea transporter lipid raft compartmentalization. FASEB J. 2011, 25, 4531–4539. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Camussi, G.; Valadi, H.; Nazarenko, I.; Ekstrom, K.; Wang, X.; Principe, S.; Shah, N.; Ashraf, N.M.; Fatima, F.; et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014, 11, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Katsuda, T.; Kosaka, N.; Ochiya, T. The roles of extracellular vesicles in cancer biology: Toward the development of novel cancer biomarkers. Proteomics 2014, 14, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I. Mucin-type O-glycans in human colon and breast cancer: Glycodynamics and functions. EMBO Rep. 2006, 7, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, J.Q.; Kruger, A.; Gallogly, S.; Hanley, S.A.; Hogan, M.C.; Ward, C.J.; Joshi, L.; Griffin, M.D. Surface glycosylation profiles of urine extracellular vesicles. PLoS ONE 2013, 8, e74801. [Google Scholar] [CrossRef] [PubMed]
- Staubach, S.; Schadewaldt, P.; Wendel, U.; Nohroudi, K.; Hanisch, F.G. Differential glycomics of epithelial membrane glycoproteins from urinary exovesicles reveals shifts toward complex-type N-glycosylation in classical galactosemia. J. Proteome Res. 2012, 11, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Runz, S.; Keller, S.; Rupp, C.; Stoeck, A.; Issa, Y.; Koensgen, D.; Mustea, A.; Sehouli, J.; Kristiansen, G.; Altevogt, P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007, 107, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Grassadonia, A.; Tinari, N.; Iurisci, I.; Piccolo, E.; Cumashi, A.; Innominato, P.; D’Egidio, M.; Natoli, C.; Piantelli, M.; Iacobelli, S. 90K (Mac-2 BP) and galectins in tumor progression and metastasis. Glycoconj. J. 2004, 19, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Borgiani, B.; Verderio, C.; Furlan, R. Microvesicles: Novel biomarkers for neurological disorders. Front. Physiol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [PubMed]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.; Hall, G.F. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Liu, C.; Cook, T.J.; Bullock, K.M.; Zhao, Y.; Ginghina, C.; Li, Y.; Aro, P.; Dator, R.; He, C.; et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014, 128, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Keller, S.; Altevogt, P.; Costa, J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 2007, 428, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Grad, L.I.; Yerbury, J.J.; Turner, B.J.; Guest, W.C.; Pokrishevsky, E.; O’Neill, M.A.; Yanai, A.; Silverman, J.M.; Zeineddine, R.; Corcoran, L.; et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 3620–3625. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Brettschneider, J.; del Tredici, K.; Lee, V.M.; Trojanowski, J.Q. Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 2015, 16, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Candelario, K.M.; Steindler, D.A. The role of extracellular vesicles in the progression of neurodegenerative disease and cancer. Trends Mol. Med. 2014, 20, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Kourembanas, S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015, 77, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014. [Google Scholar] [CrossRef] [PubMed]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Franzen, C.A.; Simms, P.E.; van Huis, A.F.; Foreman, K.E.; Kuo, P.C.; Gupta, G.N. Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Bai, O.; Li, F.; Yuan, J.; Laferte, S.; Xiang, J. Mature dendritic cells pulsed with exosomes stimulate efficient cytotoxic T-lymphocyte responses and antitumour immunity. Immunology 2007, 120, 90–102. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Maguire, C.A.; Balaj, L.; Sivaraman, S.; Crommentuijn, M.H.; Ericsson, M.; Mincheva-Nilsson, L.; Baranov, V.; Gianni, D.; Tannous, B.A.; Sena-Esteves, M.; et al. Microvesicle-associated AAV vector as a novel gene delivery system. Mol. Ther. 2012, 20, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013, 21, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.M.; Wiklander, P.B.; Nordin, J.Z.; Al-Shawi, R.; Wood, M.J.; Vithlani, M.; Schapira, A.H.; Simons, J.P.; El-Andaloussi, S.; Alvarez-Erviti, L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord. 2014, 29, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Alves, P.; Serra, M.; Brito, C.; Luis, R.B.; Lopez, J.A.; Vazquez, J.; Carrondo, M.J.; Bernad, A.; Alves, P.M. Exploring analytical proteomics platforms toward the definition of human cardiac stem cells receptome. Proteomics 2015, 15, 1332–1337. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, J.; Gomes-Alves, P.; Carvalho, S.B.; Peixoto, C.; Alves, P.M.; Altevogt, P.; Costa, J. Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures. Biomolecules 2015, 5, 1741-1761. https://doi.org/10.3390/biom5031741
Gomes J, Gomes-Alves P, Carvalho SB, Peixoto C, Alves PM, Altevogt P, Costa J. Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures. Biomolecules. 2015; 5(3):1741-1761. https://doi.org/10.3390/biom5031741
Chicago/Turabian StyleGomes, Joana, Patrícia Gomes-Alves, Sofia B. Carvalho, Cristina Peixoto, Paula M. Alves, Peter Altevogt, and Julia Costa. 2015. "Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures" Biomolecules 5, no. 3: 1741-1761. https://doi.org/10.3390/biom5031741
APA StyleGomes, J., Gomes-Alves, P., Carvalho, S. B., Peixoto, C., Alves, P. M., Altevogt, P., & Costa, J. (2015). Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures. Biomolecules, 5(3), 1741-1761. https://doi.org/10.3390/biom5031741





