The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata
Abstract
:1. Introduction
2. Material and Methods
2.1. Organisms
2.2. Growth Conditions
2.3. Antifungal Drugs
2.4. Biofilm Matrix Structure: Confocal Laser Scanning Microscopy
2.5. Biofilm Antifungal Susceptibility Tests
2.6. Effect of the Antifungals on Biofilm Cells’ Walls and on Biofilm Matrix Composition
2.6.1. Matrix Extraction Method
2.6.2. Quantification of Mannans on Biofilm Cells and on Biomass—Quantitative Alcian Blue Binding Assay
2.6.3. Alcian Blue Quantitative Assay on Biofilms
2.6.4. Isolation of Candida glabrata Biofilm Cells Walls
2.6.5. β-1,3 glucans Concentration Determination
2.7. Statistical Analysis
3. Results
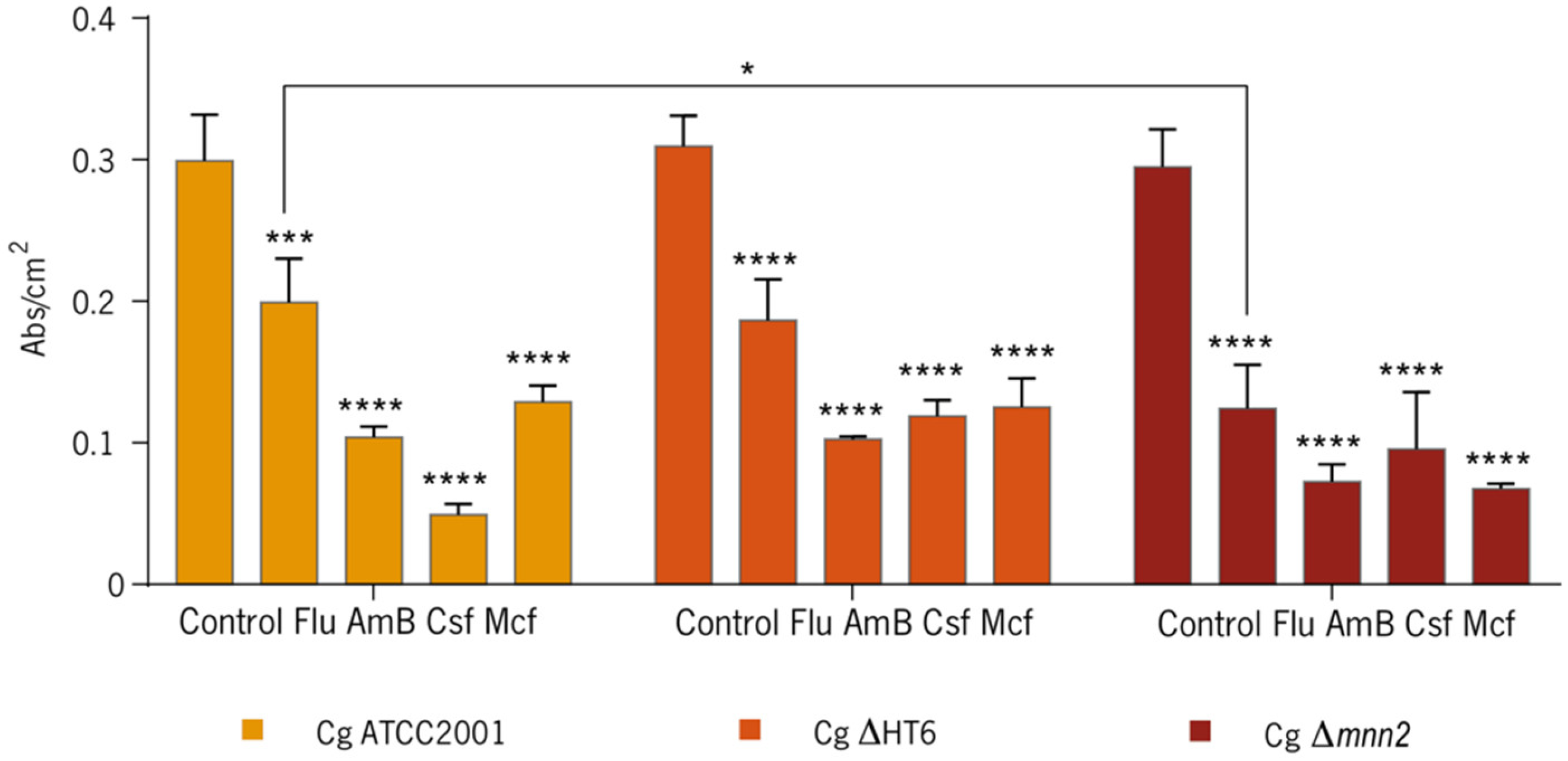
3.1. Effect of the Antifungals on Biofilms
3.2. Effect of the Antifungals on Biofilm’s Cells Walls and Matrix
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rodrigues, C.F.; Rodrigues, M.; Silva, S.; Henriques, M. Candida glabrata Biofilms: How Far Have We Come? J. Fungi 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.A.; Bates, S.; Lenardon, M.D.; Maccallum, D.M.; Wagener, J.; Lowman, D.W.; Kruppa, M.D.; Williams, D.L.; Odds, F.C.; Brown, A.J.P.; et al. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 2013, 9, e1003276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, A.L.; Guimarães, T.; Silva, L.R.; de Almeida Monfardini, L.P.; Cunha, A.K.; Rady, P.; Alves, T.; Rosas, R.C. prospective observational study of candidemia in sao paulo, brazil: incidence rate, epidemiology, and predictors of mortality. Infect. Control Hosp. Epidemiol. 2007, 28, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Xess, I.; Wang, X.; Jain, N.; Fries, B.C. Biofilm formation in clinical Candida isolates and its association with virulence. Microbes Infect. 2009, 11, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Sardi, J.C.O.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.M.; Mendes Giannini, M.J.S. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013, 62, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.S.-Y.; Rosli, R.; Seow, H.F.; Chong, P.P. Candida and invasive candidiasis: Back to basics. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.-Y.; Lee, L.-N.; Jerng, J.-S.; Yu, C.-J.; Hsueh, P.-R.; Saral, R. Candida glabrata fungaemia in intensive care units. Clin. Microbiol. Infect. 2008, 14, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.; Sardi, J.; Pitangui, N.; de Oliveira, H.; Scorzoni, L.; Galeane, M.; Medina-Alarcón, K.; Melo, W.; Marcelino, M.; Braz, J.; et al. Fungal Biofilms and Polymicrobial Diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- West, L.; Lowman, D.; Mora-Montes, H.; Grubb, S.; Murdoch, C.; Thornhill, M.; Gow, N.; Williams, D.; Haynes, K. Differential virulence of Candida glabrata glycosylation mutants. J. Biol. Chem. 2013, 288, 22006–22018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A.R. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.-P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.; Hube, B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012, 15, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Murciano, C.; Moyes, D.L.; Runglall, M.; Islam, A.; Mille, C.; Fradin, C.; Poulain, D.; Gow, N.A.R.; Naglik, J.R. The mnn2 mutant of Saccharomyces cerevisiae is affected in phosphorylation of N-linked oligosaccharides. FEBS Lett. 2000, 475, 111–116. [Google Scholar]
- Dean, N. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1999, 1426, 309–322. [Google Scholar] [CrossRef]
- Hall, R.A.; Gow, N.A.R. Mannosylation in Candida albicans: Role in cell wall function and immune recognition. Mol. Microbiol. 2013, 90, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Kudoh, A.; Okawa, Y.; Shibata, N. Significant differences in the cell-wall mannans from three Candida glabrata strains correlate with antifungal drug sensitivity. FEBS J. 2012, 279, 1844–1856. [Google Scholar] [CrossRef] [PubMed]
- Okawa, Y.; Goto, K. Disappearance of antigenic factor 6 in Candida glabrata IFO 0622 strain cells cultured at high temperature. Biol. Pharm. Bull. 2006, 29, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Diaz-Jimenez, D.F.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.P.; Odds, F.C.; et al. Endoplasmic Reticulum-Glycosidases of Candida albicans Are Required for N Glycosylation, Cell Wall Integrity, and Normal Host-Fungus Interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.H.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Brown, A.J.P.; Odds, F.C.; et al. Outer Chain N-Glycans Are Required for Cell Wall Integrity and Virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Rayner, J.C.; Munro, S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26836–26843. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.W.; Wilson, M.J.; Lewis, M.A.O.; Potts, A.J.C. Identification of Candida Species by PCR and Restriction Fragment Length Polymorphism Analysis of Intergenic Spacer Regions of Ribosomal DNA. J. Clin. Microbiol. 1995, 33, 2476–2479. [Google Scholar] [PubMed]
- Fonseca, E.; Silva, S.; Rodrigues, C.F.; Alves, C.; Azeredo, J.; Henriques, M. Effects of fluconazole on Candida glabrata biofilms and its relationship with ABC transporter gene expression. Biofouling 2014, 30, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Singleton, D.R.; Masuoka, J.; Hazen, K.C. Surface Hydrophobicity Changes of Two. Society 2005, 4, 639–648. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Victoria Elorza, M.; Valentín, E.; Sentandreu, R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006, 6, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Arikan, S.; Barchiesi, F.; Bille, J.; Dannaoui, E.; Denning, D.W.; Donnelly, J.P.; Fegeler, W.; Moore, C.; Richardson, M.; et al. EUCAST Technical Note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia—Forming moulds. Clin. Microbiol. Infect. 2008, 14, 982–984. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs, Version 8.1. Available online: http://www.eucast.org/ast_of_bacteria/previous_versions_of_documents/ (accessed on 30 October 2018).
- Rodrigues, C.F.; Henriques, M. Portrait of Matrix Gene Expression in Candida glabrata Biofilms with Stress Induced by Different Drugs. Genes 2018, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Rodrigues, M.E.; Henriques, M. Susceptibility of Candida glabrata biofilms to echinocandins: Alterations in the matrix composition. Biofouling 2018, 34, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Odani, T.; Shimma, Y.I.; Wang, X.H.; Jigami, Y. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 1997, 420, 186–190. [Google Scholar] [CrossRef]
- Pitarch, A.; Sánchez, M.; Nombela, C.; Gil, C. Sequential fractionation and two-dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteom. 2002, 1, 967–982. [Google Scholar] [CrossRef]
- Hawser, S.P.; Douglas, L.J. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 1995, 39, 2128–2131. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Rodrigues, C.F.; Araújo, D.; Rodrigues, M.; Henriques, M. Candida Species Biofilms’ Antifungal Resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Henriques, M. Liposomal and Deoxycholate Amphotericin B Formulations: Effectiveness against Biofilm Infections of Candida spp. Pathogens 2017, 6, 62. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, V.; Di Campli, E.; Fazii, P.; Traini, T.; Cellini, L.; Di Giulio, M. Candida species isolated from different body sites and their antifungal susceptibility pattern: Cross-analysis of Candida albicans and Candida glabrata biofilms. Med. Mycol. 2016, 55, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Castanheira, M. Nosocomial candidiasis: Antifungal stewardship and the importance of rapid diagnosis. Med. Mycol. 2015, 54, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Murciano, C.; Moyes, D.L.; Runglall, M.; Islam, A.; Mille, C.; Fradin, C.; Poulain, D.; Gow, N.A.R.; Naglik, J.R. Candida albicans Cell Wall Glycosylation May Be Indirectly Required for Activation of Epithelial Cell Proinflammatory Responses. Infect. Immun. 2011, 79, 4902–4911. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; Torelli, R.; Posteraro, B.; Sanglard, D. Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS ONE 2011, 6, e17589. [Google Scholar] [CrossRef] [PubMed]
- Mathé, L.; Van Dijck, P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr. Genet. 2013, 59, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: Implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L. Detection and Clinical Significance of Variability among Candida Isolates. In Candida and Candidiasis, 2nd ed.; American Society of Microbiology: Washington, DC, USA, 2012; pp. 91–99. [Google Scholar]
- Ivanova, K.; Ramon, E.; Hoyo, J.; Tzanov, T. Innovative Approaches for Controlling Clinically Relevant Biofilms: Current Trends and Future Prospects. Curr. Top. Med. Chem. 2017, 17, 1889–1914. [Google Scholar] [CrossRef]
- Debono, M.; Gordee, R.S.S. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 1994, 48, 471–497. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, N.P.; Lewis, R.E. The echinocandin antifungals: An overview of the pharmacology, spectrum and clinical efficacy. Expert Opin. Investig. Drugs 2003, 12, 1313–1333. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R. Components of the fungal cell wall that turn on and turn off the immune inflammatory response. In Proceedings of the TIMM2015, Lisbon, Portugal, 9–12 October 2015. [Google Scholar]
- Chandra, J.; Mukherjee, P.K. Candida Biofilms: Development, Architecture, and Resistance. Microbiol. Spectr. 2015, 3, 157–176. [Google Scholar] [CrossRef] [PubMed]




| Strain | Control | Flu | AmB | Csf | Mcf |
|---|---|---|---|---|---|
| C. glabrata ATCC2001 | 5.90 ± 0.18 | 4.95 ± 0.44 | 4.40 ± 0.30 | 3.80 ± 1.27 | 3.40 ± 0.07 |
| C. glabrata ΔHT6 | 5.80 ± 0.36 | 3.69 ± 0.02 | 4.37 ± 0.45 | 5.42 ± 0.42 | 5.53 ± 0.41 |
| C. glabrata Δmnn2 | 6.16 ± 0.31 | 5.10 ± 0.43 | 4.23 ± 0.58 | 5.61 ± 0.11 | 4.98 ± 0.13 |
| Strain | β-1,3 glucans Biofilm Cells Concentration (pg/mL) ± SD |
|---|---|
| C. glabrata ATCC2001 | 359.00 ± 8.20 |
| C. glabrata ΔHT6 | 370.00 ± 5.08 |
| C. glabrata Δmnn2 | 387.00 ± 7.00 (*) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, C.F.; Vilas Boas, D.; Haynes, K.; Henriques, M. The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata. Biomolecules 2018, 8, 130. https://doi.org/10.3390/biom8040130
Rodrigues CF, Vilas Boas D, Haynes K, Henriques M. The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata. Biomolecules. 2018; 8(4):130. https://doi.org/10.3390/biom8040130
Chicago/Turabian StyleRodrigues, Célia F., Diana Vilas Boas, Ken Haynes, and Mariana Henriques. 2018. "The MNN2 Gene Knockout Modulates the Antifungal Resistance of Biofilms of Candida glabrata" Biomolecules 8, no. 4: 130. https://doi.org/10.3390/biom8040130






