Exploring the Antioxidant Effects and Periodic Regulation of Cancer Cells by Polyphenols Produced by the Fermentation of Grape Skin by Lactobacillus plantarum KFY02
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fermentation
2.2. In Vitro Antioxidant Analysis
2.3. Culture of Cell
2.4. Oxidative Damage Model
2.5. Apoptosis Model
2.6. Flow Cytometry
2.7. RNA Extraction and qRT-PCR
2.8. Protein Isolation and Western Blotting Analysis
2.9. High-Performance Liquid Chromatography (HPLC)
2.10. Statistical Analysis
3. Results
3.1. Antioxidant Activity In Vitro
3.2. H2O2-Induced Oxidative Damage and Effects of KFSE on Human Embryonic Kidney (293T) Cell
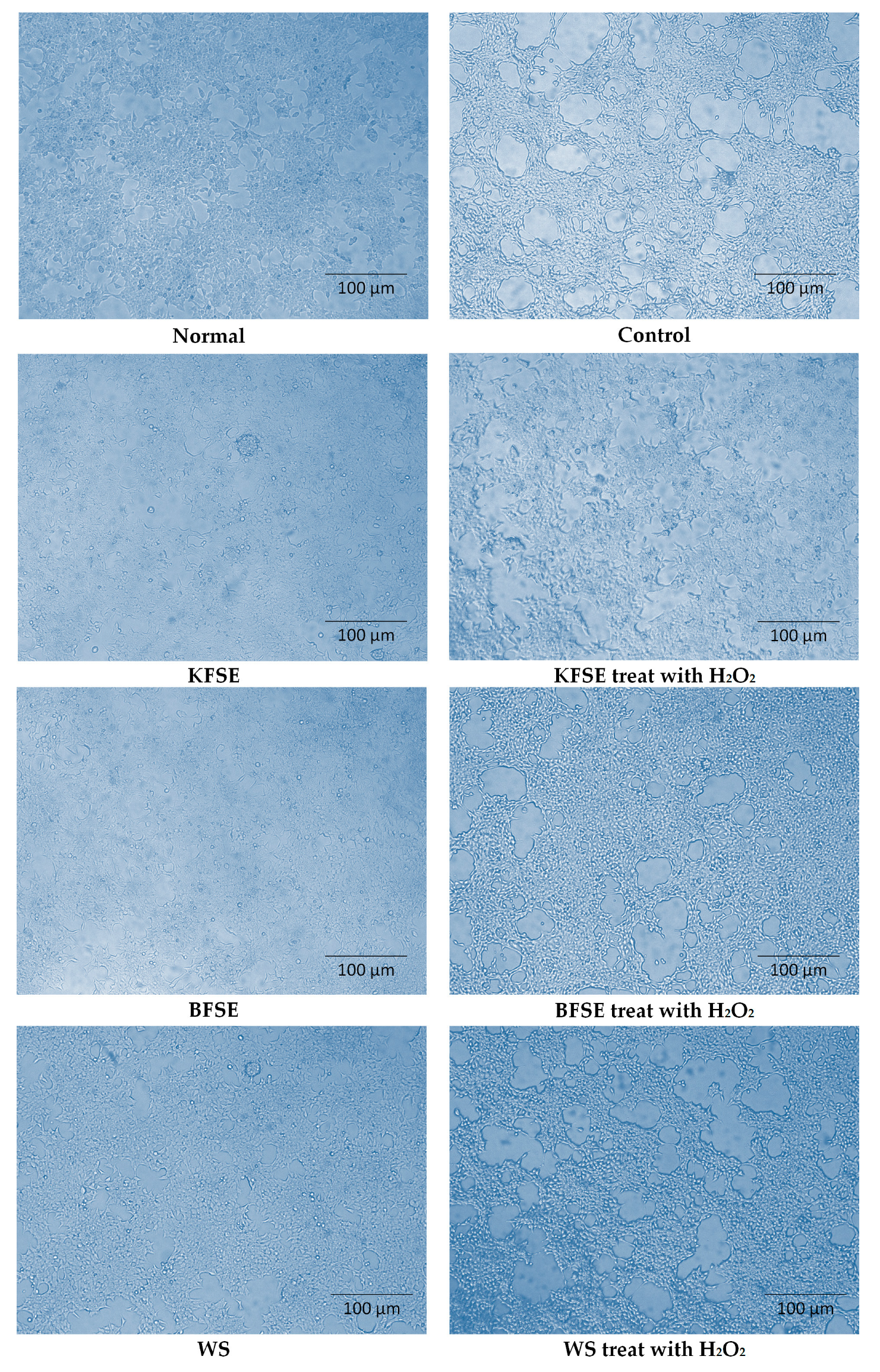
3.3. Effect of KFSE on Human Embryonic Kidney (293T) Cell Morphology
3.4. Oxidation Index of the Human Embryonic Kidney (293T) Cell
3.5. KFSE Promotes SOD, CAT, GSH, and GSH-Px mRNA and Protein Expression in the Human Embryonic Kidney (293T) Cell
3.6. KFSE Inhibits Human Hepatoma (HepG2) Cells Growth In Vitro
3.7. KFSE Induces Cell Cycle Arrest in Human Hepatoma (HepG2) Cells
3.8. Protein Expression and mRNA Expression in Human Hepatoma (HepG2) Cells
3.9. Constituents of Fermentation Solution
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Gene | Primer Sequence | X (Annealing Temperature (°C)) |
|---|---|---|
| GAPDH | F: 5′-TCAAGAAGGTGGTGAAGCAGG-3′ R: 5′-AGCGTCAAAGGTGGAGGAGTG-3′ | |
| CAT | F: 5′-TGGAGCTGGTAACCCAGTAGG-3′ R: 5′-CCTTTGCCTTGGAGTATTTGGTA-3′ | 57.0 |
| SOD1 | F: 5′-GGTGGGCCAAAGGATGAAGAG-3′ R: 5′-CCACAAGCCAAACGACTTCC-3′ | 57.0 |
| GSH | F: 5′-GGGAGCCTCTTGCAGGATAAA-3′ R: 5′-GAATGGGGCATAGCTCACCAC-3′ | 57.0 |
| GSH-Px | F: 5′-CAGTCGGTGTATGCCTTCTCG-3′ R: 5′-GAGGGACGCCACATTCTCG-3′ | 57.0 |
| p53 | F: 5′-CTTTGAGGTGCGTGTTTGTGC-3′ R: 5′-GGTTTCTTCTTTGGCTGGGGA-3′ | 51.6 |
| CDK4 | F: 5′-ATGGCTACCTCTCGATATGAGC-3′ R: 5′-CATTGGGGACTCTCACACTCT-3′ | 56.6 |
| Cyclin-D1 | F: 5′-GCTGCGAAGTGGAAACCATC-3′ R: 5′-CCTCCTTCTGCACACATTTGAA-3′ | 60.3 |
| p21 | F: 5′-TGTCCGTCAGAACCCATGC-3′ R: 5′-AAAGTCGAAGTTCCATCGCTC-3′ | 54.9 |
| pRb1 | F: 5′-CTCTCGTCAGGCTTGAGTTTG-3′ R: 5′GACATCTCATCTAGGTCAACTGC-3′ | 66.0 |
| C-myc | F: 5′-TGGAACGTCAGAGGAGAAACGA-3′ R: 5′-CTTGAACGGACAGGATGTAGGC-3′ | 50.0 |
| Caspase-3 | F: 5′-CATGGAAGCGAATCAATGGACT-3′ R: 5′-CTGTACCAGACCGAGATGTCA-3′ | 50.7 |
| Caspase-8 | F: 5′-ATTTTGAGATCAAGCCCCACG-3′ R: 5′-GGATACAGCAGATGAAGCAGTCC-3′ | 66.0 |
| Caspase-9 | F: 5′-CTCAGACCAGAGATTCGCAAAC-3′ R: 5′-GCATTTCCCCTCAAACTCTCAA-3′ | 50.0 |
| Caspase-7 | F: 5′-CGGTCCTCGTTTGTACCGTC-3′ R: 5′-CGCCCATACCTGTCACTTTATCA-3′ | 64.4 |
| cox-2 | F: 5′-CTGGCGCTCAGCCATACAG-3′ R: 5′-CGCACTTATACTGGTCAAATCCC-3′ | 65.0 |
| Bcl-2 | F: 5′-ATGTGTGTGGAGAGCGTCAACC-3′ R: 5′-CAGAGACAGCCAGGAGAAATCAA-3′ | 65.0 |
| NF-κB | F: 5′-GAAGCACGAATGACAGAGGC-3′ R: 5′-GCTTGGCGGATTAGCTCTTTT-3′ | 50.9 |
| PCNA | F: 5′-CCAGGGCTCCATCCTCAAGAA-3′ R: 5′-GACGTGGGACGAGTCCATGCT-3′ | 62.0 |
| TGF-β | F: 5′-CAGCGTGCCTAAACTTTATCAGC-3′ R: 5′-TCAGGAGGATGTTTCACATGGA-3′ | 58.2 |
References
- Bosch, F.X.; Ribes, J.; Mireia, D.; Ramon, C. Primary liver cancer: Worldwide incidence and trends. Gastroenterology 2004, 127, S5–S16. [Google Scholar] [CrossRef]
- Valery, P.C.; Laversanne, M.; Clark, P.J.; Petrick, J.L.; Mcglynn, K.A.; Bray, F. Projections of primary hepatoma to 2030 in 30 countries worldwide. Hepatology 2018, 67, 600–611. [Google Scholar] [CrossRef]
- Gay, N.H.; Phopin, K.; Suwanjang, W.; Songtawee, N.; Ruankham, W.; Wongchitrat, P.; Prachayasittikul, S.; Prachayasittikul, V. Neuroprotective effects of phenolic and carboxylic acids on oxidative stress-induced toxicity in human neuroblastoma SH-SY5Y cells. Neurochem. Res. 2018, 43, 1–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, Y.; Wu, L.; Meng, Q.; Liu, M.Z. Effect of resveratrol on proliferation and autophagy of human hepatoma HepG2 cells. Chin. J. Biol. 2018, 31, 607–612. [Google Scholar]
- Rush, E.; Ferguson, L.R.; Cumin, M.; Thakur, V.; Karunasinghe, N.; Plank, L. Kiwifruit consumption reduces DNA fragility: A randomized controlled pilot study in volunteers. Nutr. Res. 2006, 26, 197–201. [Google Scholar] [CrossRef]
- Giampieri, F.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Mazzoni, L.; Capocasa, F.; Sabbadini, S.; Alvarez-Suarez, J.M.; Afrin, S.; Rosati, C.; Pandolfini, T. Overexpression of the anthocyanidin synthase gene in strawberry enhances antioxidant capacity and cytotoxic effects on human hepatic cancer cells. J. Agric. Food Chem. 2018, 66, 581–592. [Google Scholar] [CrossRef]
- Saaara, Y.; Miyata, Y.; Nakamura, Y.; Yasuda, T.; Araki, K.; Matsuo, T.; Ohba, K.; Sakai, H. Anti-cancer effects of green tea consumption on malignant behavior up-regulated by smoking in patients with upper urinary tract cancer. Eur. Urol. Suppl. 2018, 17, e1439. [Google Scholar] [CrossRef]
- Kammerer, D.; Claus, A.; Carle, R.; Schieber, A. Polyphenol screening of pomace from red and white grape varieties (Vitis vinifera L.) by HPLC-DAD-MS/MS. J. Agric. Food Chem. 2004, 52, 4360–4367. [Google Scholar] [CrossRef]
- Eremina, A.V.; Reshetnyak, V.Y.; Vezirishvili, M.O. HPLC determination of polyphenols in dry grape stem extract. Pharm. Chem. J. 2004, 38, 143–145. [Google Scholar] [CrossRef]
- Scalbert, A.; Johnson, I.T.; Saltmarsh, M. Polyphenols: Antioxidants and beyond. Am. J. Clin. Nutr. 2005, 81, 215S–217S. [Google Scholar] [CrossRef]
- Kim, S.; Bang, H.; Yoo, K.S.; Pike, L.M. Overexpression of Anthocyanidin Synthase (ANS) gene in strawberry enhances antioxidant capacity and cytotoxic effects on human hepatic cancer cells. J. Agric. Euphytica. 2006, 145, 45–51. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Identification of the fourth allele of the ans (anthocyanidin synthase) gene and its effect on red color intensity in onions (allium cepa). Gut Pathog. 2018, 10, 21–40. [Google Scholar] [CrossRef]
- Shori, A.B. Microencapsulation improved probiotics survival during gastric transit. Hayati J. Biosci. 2018, 24, 1–5. [Google Scholar] [CrossRef]
- Swati, S.M.; Prafulla, K.B.; Biswabandita, K.; Ramesh, C.R. Advances in probiotics, prebiotics and nutraceuticals. Innov. Technol. Fermented Food Beverage Ind. 2018, 5, 121–141. [Google Scholar]
- George, K.R.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Azcárate-Peril, M.A.; Sikes, M.; Bruno-Bárcena, J.M. The intestinal microbiota, gastrointestinal environment and colorectal cancer: A putative role for probiotics in prevention of colorectal cancer? Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G401–G424. [Google Scholar]
- Bin, X.; Chen, J.; Zhou, Y. Cellular oxidative damage of HEK293T cells induced by combination of CdCl2 and Nano-TiO2. J. Huazhong Univ. Sci. Technol. Med. Sci. 2011, 31, 290–294. [Google Scholar]
- Thaipong, K.; Boonprakob, U.; Crosby, K.; Cisneros, Z.L.; Byrne, D.H. Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J. Food Compos. Anal. 2006, 19, 669–675. [Google Scholar] [CrossRef]
- Bernini, R.; Barontini, M.; Cis, V.; Carastro, I.; Tofani, D.; Chiodo, R.A.; Lupattelli, P.; Incerpi, S. Synthesis and evaluation of the antioxidant activity of lipophilic phenethyl trifluoroacetate esters by in vitro ABTS, DPPH and in cell-culture DCF assays. Molecules 2018, 23, 208. [Google Scholar] [CrossRef]
- Akalin, A.; Elmore, L.W.; Forsythe, H.L.; Amaker, B.A.; Holt, S.E. A novel mechanism for chaperone-mediated telomerase regulation during prostate cancer progression. Cancer Res. 2001, 61, 4791–4796. [Google Scholar]
- Juan, M.E.; Wenzel, U.; Daniel, H.; Planas, J.M. Resveratrol induces apoptosis through ROS-dependent mitochondria pathway in HT-29 human colorectal carcinoma cells. J. Agric. Food Chem. 2008, 56, 4813–4818. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.K.; Lee, E.J.; Kim, H.C.; Kim, J.H. Induction of G(1)/S phase arrest and apoptosis by quercetin in human osteosarcoma cells. Arch. Pharm. Res. 2010, 33, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Ansari, S.H.; Nazish, I. Bio-functional aspects of grape seeds—A review. Int. J. Phytomed. 2011, 2, 177–181. [Google Scholar]
- Nardini, M.; Garaguso, I. Effect of sulfites on antioxidant activity, total polyphenols, and flavonoid measurements in white wine. Foods 2018, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, H.; Sakagami, H.; Hatano, T. Three new flavonoids, proanthocyanidin, and accompanying phenolic constituents from Feijoa sellowiana. Biosci. Biotech. Biochem. 2018, 82, 1–11. [Google Scholar]
- Lili, X.U.; Yue, Q.Y.; Bian, F.E.; Zhai, H.; Yao, Y.X. Melatonin Treatment enhances the polyphenol content and antioxidant capacity of red wine. Hortic. Plant J. 2018, 4, 144–150. [Google Scholar]
- Tzanova, M.; Peeva, P. Rapid HPLC method for simultaneous quantification of trans-resveratrol and quercetin in the skin of red grapes. Food Anal. Methods 2018, 11, 514–521. [Google Scholar] [CrossRef]
- Xue, B.; Lu, Q.Y.; Massie, L.; Qualls, C.; Mao, J.T. Grape seed procyanidin extract against lung cancer: The role of microrna-106b, bioavailability, and bioactivity. Oncotarget 2018, 9, 15579–15590. [Google Scholar] [CrossRef]
- Eng, E.T.; Ye, J.D.; Phung, S.; Moore, R.E.; Young, M.K.; Gruntmanis, U.; Braunstein, G.; Chen, S. Suppression of estrogen biosynthesis by procyanidin dimers in red wine and grape seeds. Cancer Res. 2003, 63, 8516–8522. [Google Scholar]
- Shaik, Y.B.; Castellani, M.L.; Perrella, A.; Conti, F.; Cerulli, G. Role of quercetin (a natural herbal compound) in allergy and inflammation. J. Biol. Reg. Homeos. Agents 2006, 20, 47–52. [Google Scholar]
- Shelly, H.; Corene, C.; Sun, S.; Sun, X.X.; Hoda, K.; Zhou, K.Q. Dietary supplementation of grape skin extract improves glycemia and inflammation in diet-induced obese mice fed a western high fat diet. J. Agric. Food Chem. 2011, 59, 3035–3041. [Google Scholar]
- Haddad, J.J.; Harb, H.L. L-gamma-Glutamyl-L-cysteinyl-glycine (glutathione; GSH) and GSH-related enzymes in the regulation of pro- and anti-inflammatory cytokines: A signaling transcriptional scenario for redox(y) immunologic sensor(s)? Mol. Immunol. 2005, 42, 987–1014. [Google Scholar] [CrossRef] [PubMed]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Chu, H.; Low, P.S. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. Biochem. J. 2006, 400, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Long, X.Y.; Pan, Y.N.; Zhao, X. Prophylactic effect of Kudingcha polyphenols on oxazolone induced colitis through its antioxidant capacities. Food Sci. Hum. Wellness 2018, 7, 209–214. [Google Scholar] [CrossRef]
- Kim, M.P.; Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.M.; Burke, R.T.; Doak, A.E.; Soyeon, P.; Orth, J.D. Loss of p53 expression in cancer cells alters cell cycle response after inhibition of exportin-1 but does not prevent cell death. Cell Cycle 2018, 17, 1–16. [Google Scholar] [CrossRef]
- Collins, A.R.; Ma, A.G.; Duthie, S.J. Implication of p53 in base excision DNA repair: In vivo evidence. Oncogene 2002, 21, 731–737. [Google Scholar]
- Wang, W.; Zeng, C.; Feng, Y.; Zhou, F.; Liao, F.; Liu, Y. The size-dependent effects of silica nanoparticles on endothelial cell apoptosis through activating the p53-caspase pathway. Environ. Pollut. 2018, 233, 218–225. [Google Scholar] [CrossRef]
- Zeng, L.; Xiao, Q.; Margariti, A.; Xu, Q. Shear stress-induced stem cell differentiation toward endothelial cells is regulated by HDAC-P53-P21 pathways. Vasc. Pharm. 2006, 45, e52. [Google Scholar] [CrossRef]
- Meyer, C.A.; Jacobs, H.W.; Datar, S.A.; Du, W.; Lehner, C.F. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. Embo J. 2014, 19, 4533–4542. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, R.; Nayebi, A.M.; Eyvari, S.B. Silymarin prevents apoptosis through inhibiting the Bax/caspase-3 expression and suppresses toll like receptor-4 pathway in the SNc of 6-OHDA intoxicated rats. Biomed. Pharm. 2018, 104, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Ballestri, S.; Nascimbeni, F.; Romagnoli, D.; Baldelli, E.; Lonardo, A. The role of nuclear receptors in the pathophysiology, natural course, and drug treatment of NAFLD in humans. Adv. Ther. 2016, 33, 291–319. [Google Scholar] [CrossRef] [PubMed]







| t/min | A% | B% |
|---|---|---|
| 0–20 | 97–95 | 3–5 |
| 20–35 | 95–85 | 5–15 |
| 35–50 | 85–70 | 15–30 |
| 50–65 | 70–70 | 30–30 |
| 65–75 | 70–0 | 30–0 |
| Group | LDH (U/104) | GSH (μg/104) | GSH-Px (U/mg prot) | CAT (U/104) | MDA (nmol/104) | SOD (U/104) | T-AOC (U/104) | NO (μmol/104) |
|---|---|---|---|---|---|---|---|---|
| Normal | 5.44 ± 0.16 a | 3.13 ± 0.14 c | 2.50 ± 0.20 c | 2.77 ± 0.21 c | 1.42 ± 0.04 a | 18.12 ± 0.61 d | 18.32 ± 0.79 d | 4.20 ± 0.39 a |
| Control | 8.90 ± 0.33 c | 1.89 ± 0.07 a | 0.91 ± 0.11 a | 1.43 ± 0.10 a | 2.72 ± 0.17 c | 10.46 ± 0.57 a | 11.42 ± 0.16 a | 8.43 ± 0.49 c |
| KFSE | 6.56 ± 0.64 b | 2.72 ± 0.07 b | 1.80 ± 0.07 b | 2.06 ± 0.09 b | 1.68 ± 0.06 b | 16.99 ± 0.19 c | 15.92 ± 0.25 c | 5.62 ± 0.12 b |
| BFSE | 7.40 ± 0.45 b | 2.76 ± 0.27 b | 1.62 ± 0.08 b | 1.95 ± 0.11 b | 1.75 ± 0.09 b | 16.63 ± 0.32 c | 14.62 ± 1.22 b | 4.38 ± 0.66 a |
| WS | 7.41 ± 0.40 b | 2.67 ± 0.24 b | 1.72 ± 0.03 b | 1.88 ± 0.05 b | 1.79 ± 0.08 b | 15.73 ± 0.47 b | 13.40 ± 0.31 b | 5.29 ± 0.25 b |
| Group | CAT | SOD | GSH | GSH-Px |
|---|---|---|---|---|
| Normal | 1.01 ± 0.18 c | 1.00 ± 0.09 d | 1.00 ± 0.09 c | 1.01 ± 0.18 c |
| Control | 0.39 ± 0.08 a | 0.12 ± 0.01 a | 0.32 ± 0.03 a | 0.35 ± 0.04 a |
| KFSE | 1.11 ± 0.08 c | 0.57 ± 0.09 c | 1.07 ± 0.13 c | 1.22 ± 0.08 c |
| BFSE | 0.66 ± 0.13 b | 0.30 ± 0.03 b | 0.97 ± 0.08 c | 0.72 ± 0.14 b |
| WS | 0.66 ± 0.04 b | 0.28 ± 0.08 b | 0.81 ± 0.06 b | 0.54 ± 0.16 a,b |
| Group | Bcl-2 | cox-2 | Caspase-3 | Caspase-7 | Caspase-8 |
|---|---|---|---|---|---|
| Normal | 1.01 ± 0.16 c | 1.01 ± 0.13 b | 1.01 ± 0.13 a | 1.00 ± 0.05 a | 1.13 ± 0.61 a |
| KFSE | 0.60 ± 0.08 a ↓ | 0.72 ± 0.10 a ↓ | 2.25 ± 0.56 b ↑ | 3.39 ± 0.39 c ↑ | 3.52 ± 1.07 b ↑ |
| BFSE | 0.92 ± 0.11b c ↓ | 0.76 ± 0.03 a ↓ | 1.90 ± 0.37 b ↑ | 1.14 ± 0.18 a ↑ | 2.57 ± 1.87 a,b ↑ |
| WS | 0.81 ± 0.07 b ↓ | 0.76 ± 0.04 a ↓ | 1.77 ± 0.08 b ↑ | 2.57 ± 0.88 b ↑ | 1.43 ± 0.13 a ↑ |
| Group | TGF-β | Caspase-9 | C-myc | cyclin-D1 | CDK4 |
|---|---|---|---|---|---|
| Normal | 1.14 ± 0.24 a | 1.00 ± 0.10 a | 1.00 ± 0.10 a | 1.00 ± 0.11 c | 1.00 ± 0.10 d |
| KFSE | 3.18 ± 0.17 c ↑ | 2.52 ± 0.57 d ↑ | 1.19 ± 0.13 a ↑ | 0.22 ± 0.05 a ↓ | 0.31 ± 0.04 a ↓ |
| BFSE | 5.41 ± 0.87 c ↑ | 1.39 ± 0.39 b ↑ | 1.11 ± 0.24 a ↑ | 0.36 ± 0.03 b ↓ | 0.60 ± 0.06 c ↓ |
| WS | 3.71 ± 1.07 b ↑ | 1.92 ± 0.18 c ↑ | 1.17 ± 0.17 a ↑ | 0.19 ± 0.12 a ↓ | 0.49 ± 0.04 b ↓ |
| Group | NF-κB | p21 | p53 | PCNA | qRB1 |
|---|---|---|---|---|---|
| Normal | 0.91 ± 0.24 b | 0.90 ± 0.26 a | 0.89 ± 0.34 a | 1.00 ± 0.06 b | 1.00 ± 0.05 c |
| KFSE | 0.58 ± 0.08 a ↓ | 4.48 ± 0.30 c ↑ | 2.27 ± 0.25 b ↑ | 0.18 ± 0.03 a ↓ | 0.15 ± 0.05 a ↓ |
| BFSE | 0.63 ± 0.09 a ↓ | 1.06 ± 0.12 a ↑ | 1.18 ± 0.30 a ↑ | 0.19 ± 0.01 a ↓ | 0.13 ± 0.02 a ↓ |
| WS | 0.57 ± 0.11 a ↓ | 2.66 ± 0.06 b ↑ | 1.97 ± 0.18 b ↑ | 0.21 ± 0.01 a ↓ | 0.26 ± 0.05 b ↓ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Tan, F.; Liu, X.; Yi, R.; Zhao, X. Exploring the Antioxidant Effects and Periodic Regulation of Cancer Cells by Polyphenols Produced by the Fermentation of Grape Skin by Lactobacillus plantarum KFY02. Biomolecules 2019, 9, 575. https://doi.org/10.3390/biom9100575
Liu J, Tan F, Liu X, Yi R, Zhao X. Exploring the Antioxidant Effects and Periodic Regulation of Cancer Cells by Polyphenols Produced by the Fermentation of Grape Skin by Lactobacillus plantarum KFY02. Biomolecules. 2019; 9(10):575. https://doi.org/10.3390/biom9100575
Chicago/Turabian StyleLiu, Jia, Fang Tan, Xinhong Liu, Ruokun Yi, and Xin Zhao. 2019. "Exploring the Antioxidant Effects and Periodic Regulation of Cancer Cells by Polyphenols Produced by the Fermentation of Grape Skin by Lactobacillus plantarum KFY02" Biomolecules 9, no. 10: 575. https://doi.org/10.3390/biom9100575





