Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum
Abstract
1. Introduction
2. Materials and Methods
2.1. PSIA Assay
2.2. Plasmids
2.3. Analysis of the YFP-Fused Protein Aggregation in Yeast Cells
2.4. C-DAG Assay
2.5. Protein Production, Purification, and In Vitro Fibril Formation
2.6. Transmission Electron Microscopy (TEM)
2.7. ThT staining and Confocal Microscopy
2.8. Absorbtion and Fluorescence Measurements
2.9. CR Staining and Polarization Microscopy of Fibrils
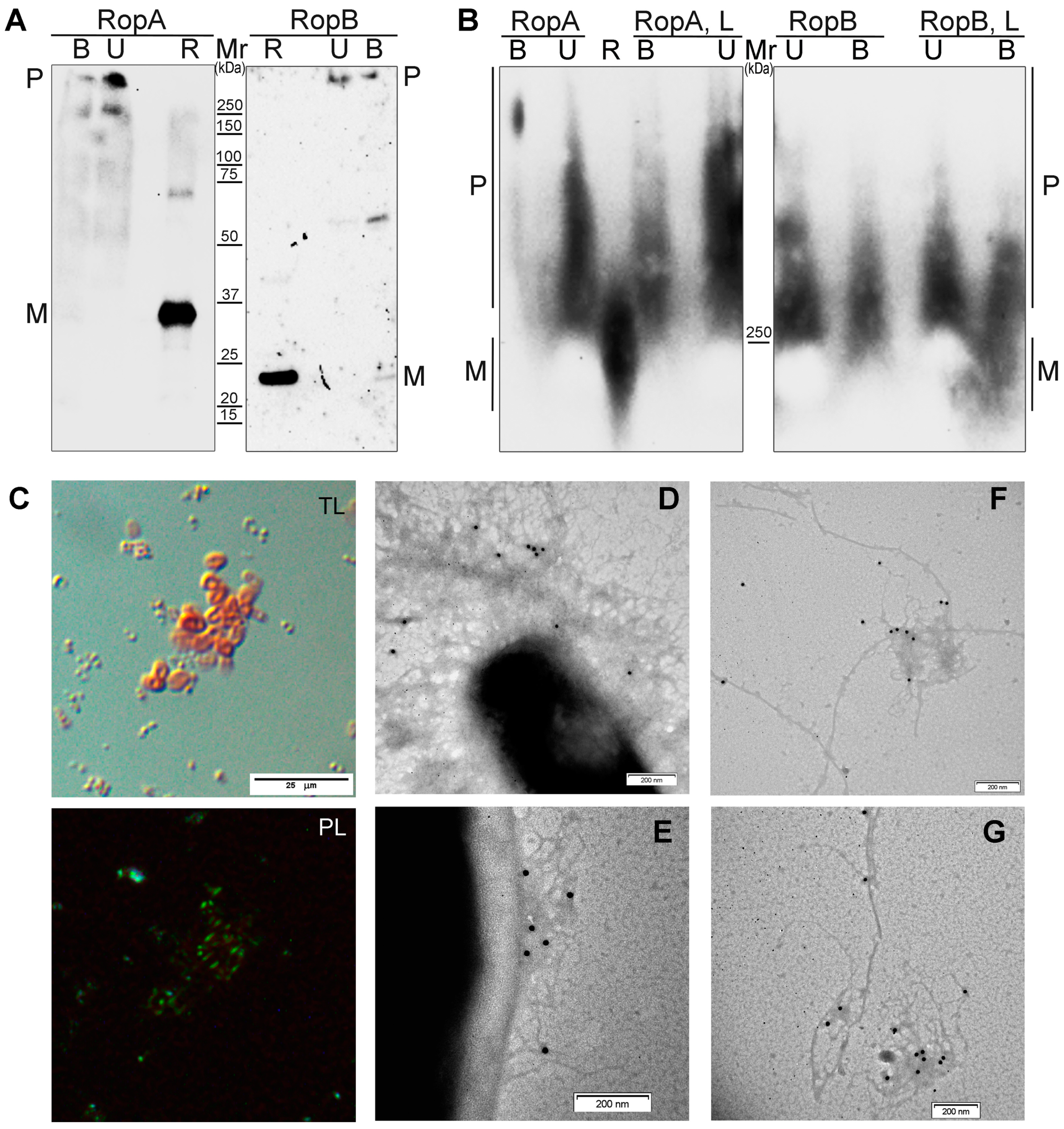
2.10. Analysis of Detergent and Protease Resistance of Protein Aggregates and Immunodetection
2.11. Data Analysis
3. Results
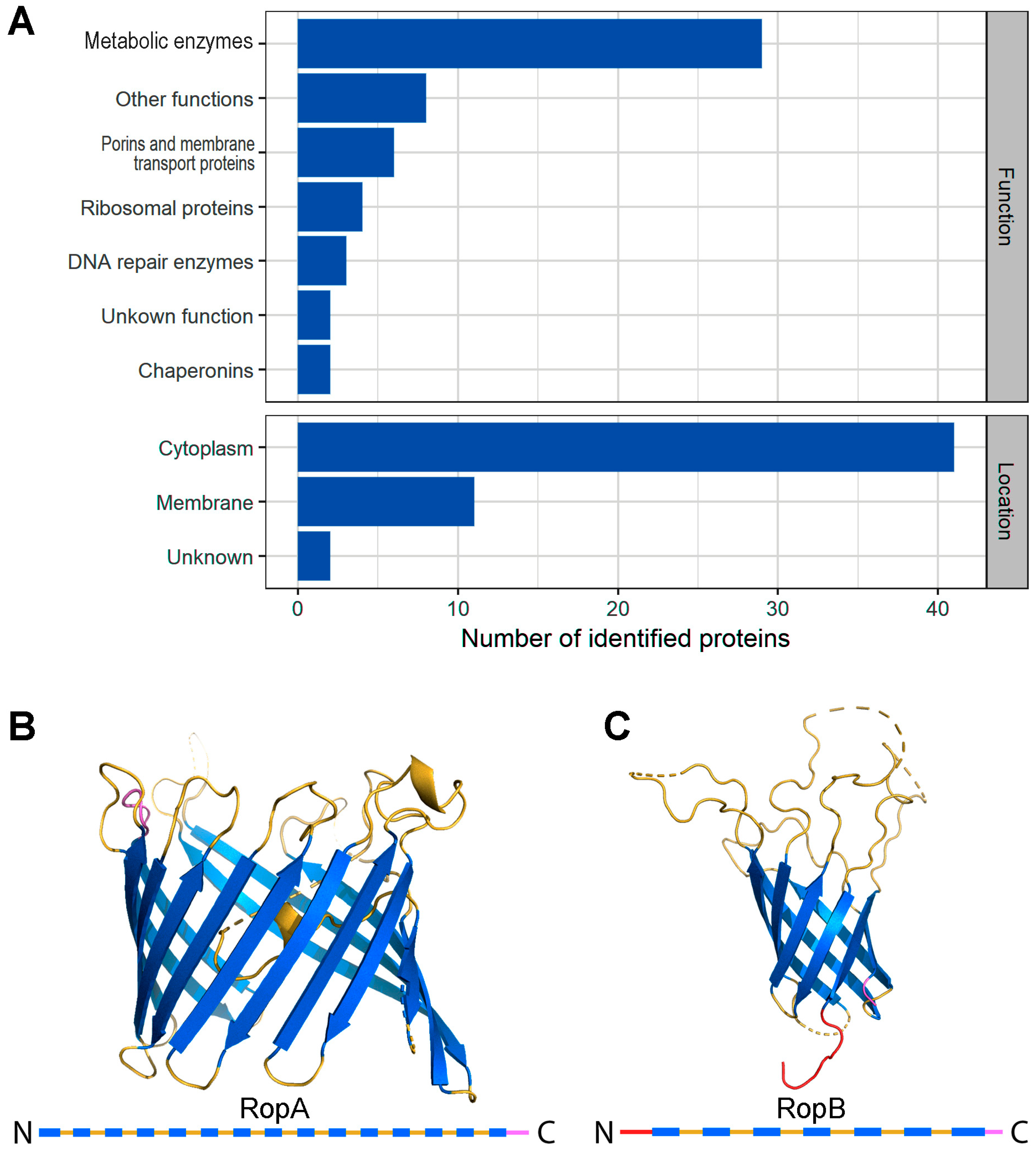
3.1. Proteomic Screening Revealed Potentially Amyloidogenic Proteins of R. leguminosarum
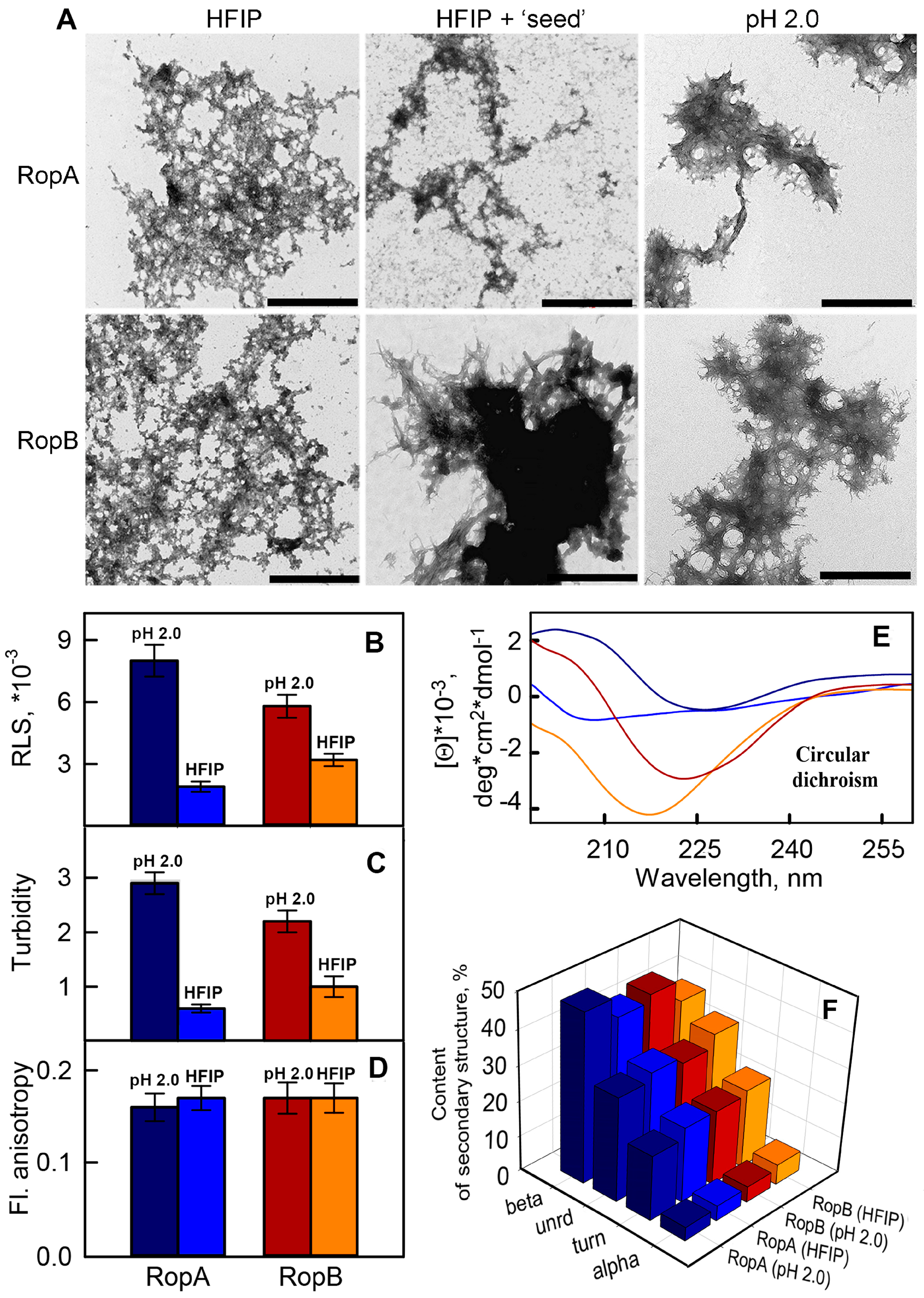
3.2. Full-Length RopA and RopB Form Fibrils In Vitro
3.3. In Vitro Obtained RopA and RopB Fibrils Exhibit Amyloid Properties
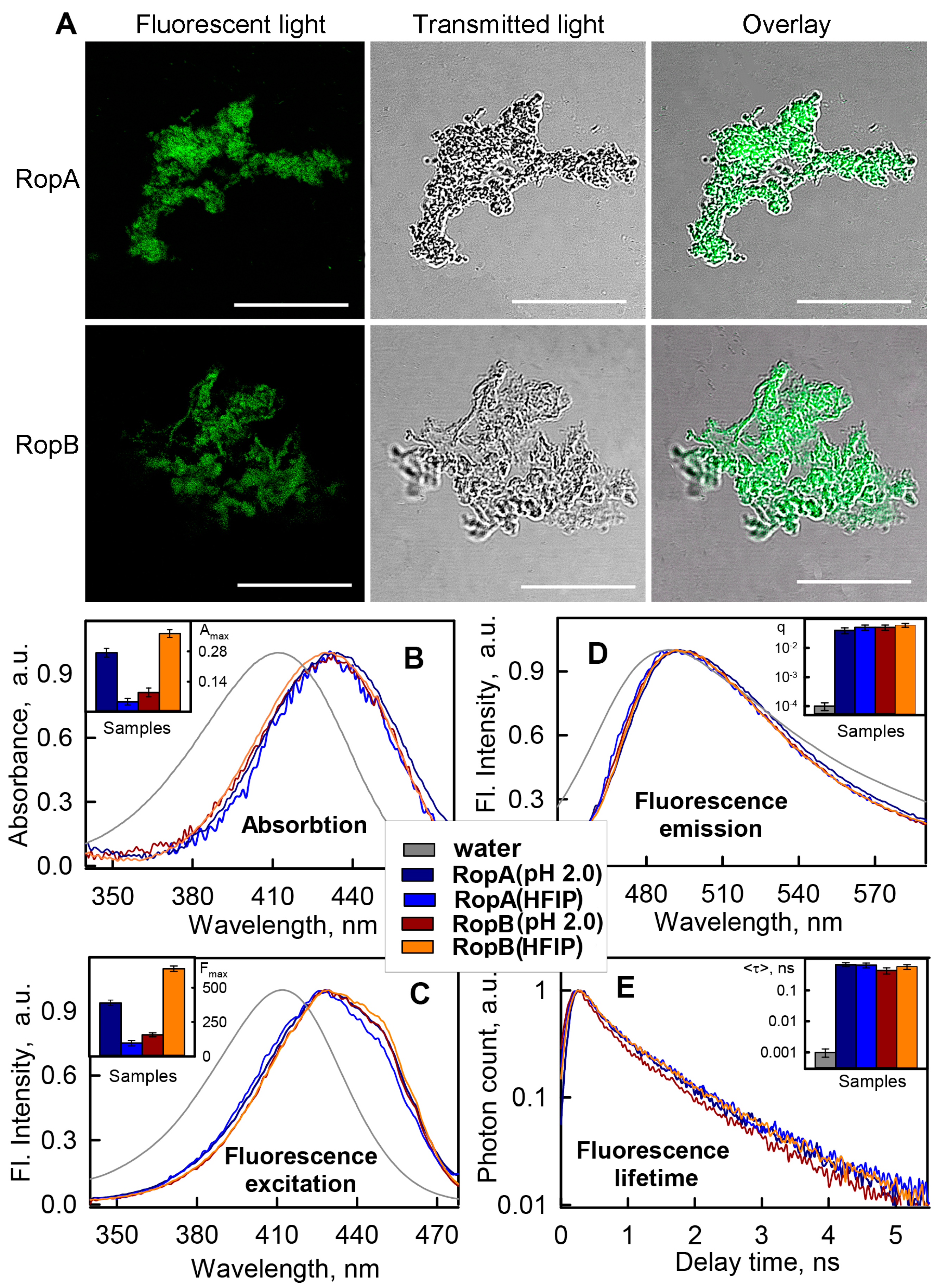
3.3.1. Photophysical Properties of the RopA and RopB Fibrils and Aggregates
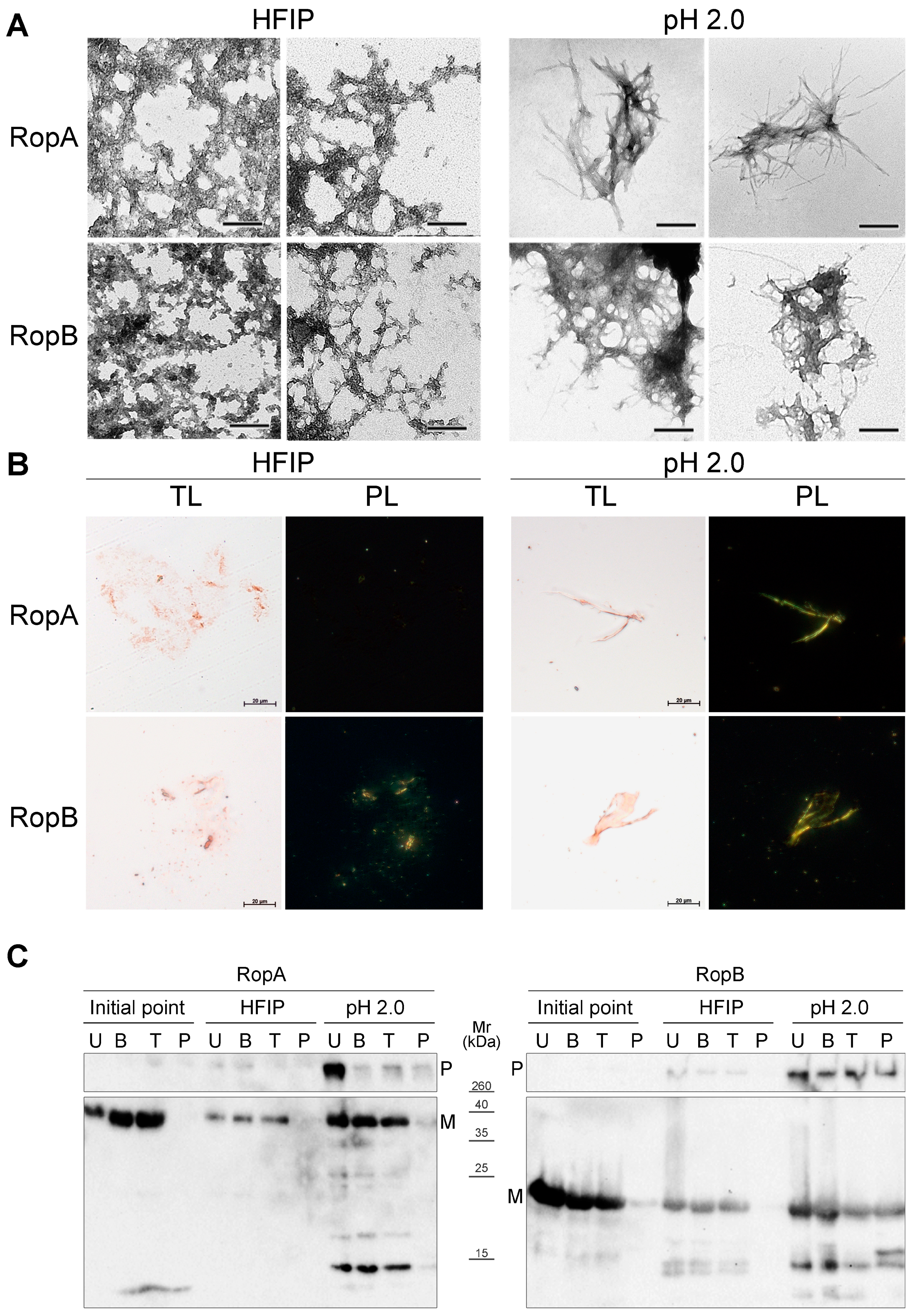
3.3.2. Fibrils of RopA and RopB Bind ThT
3.3.3. Fibrils of RopA and RopB Exhibit Green Birefringence upon Binding with CR
3.3.4. Detergent and Protease Resistance of Amyloids
3.4. RopA and RopB Demonstrate Amyloid Properties In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sipe, J.D.; Benson, M.D.; Buxbaum, J.N.; Ikeda, S.; Merlini, G.; Saraiva, M.J.M.; Westermark, P. Amyloid fibril proteins and amyloidosis: Chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid 2016, 23, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Eanes, E.D.; Glenner, G.G. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968, 16, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Sunde, M.; Serpell, L.C.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J.; Ihara, Y.; Salazar, F.J. Alzheimer’s disease: Insolubility of partially purified paired helical filaments in sodium dodecyl sulfate and urea. Science 1982, 215, 1243–1245. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.C.; McKinley, M.P.; Prusiner, S.B. Identification of a protein that purifies with the scrapie prion. Science 1982, 218, 1309–1311. [Google Scholar] [CrossRef]
- Naiki, H.; Higuchi, K.; Hosokawa, M.; Takeda, T. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavine T. Anal. Biochem. 1989, 177, 244–249. [Google Scholar] [CrossRef]
- Howie, A.J.; Brewer, D.B. Optical properties of amyloid stained by Congo red: History and mechanisms. Micron 2009, 40, 285–301. [Google Scholar] [CrossRef]
- Coustou, V.; Deleu, C.; Saupe, S.; Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 1997, 94, 9773–9778. [Google Scholar] [CrossRef]
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2017, 82, e00035-17. [Google Scholar] [CrossRef]
- Linder, M.B.; Szilvay, G.R.; Nakari-Setälä, T.; Penttilä, M.E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef]
- Holmes, D.L.; Lancaster, A.K.; Lindquist, S.; Halfmann, R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell 2013, 153, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Giustetto, M.; Etkin, A.; Hsu, R.; Janisiewicz, A.M.; Miniaci, M.C.; Kim, J.-H.; Zhu, H.; Kandel, E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 2003, 115, 893–904. [Google Scholar] [CrossRef]
- Stephan, J.S.; Fioriti, L.; Lamba, N.; Colnaghi, L.; Karl, K.; Derkatch, I.L.; Kandel, E.R. The CPEB3 Protein Is a Functional Prion That Interacts with the Actin Cytoskeleton. Cell Rep. 2015, 11, 1772–1785. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.; Cesario, W.C.; White-Grindley, E.; Jiang, H.; Ren, F.; Khan, M.R.; Li, L.; Choi, E.M.L.; Kannan, K.; Guo, F.; et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 2012, 148, 515–529. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006, 4, 100–107. [Google Scholar] [CrossRef]
- Chimileski, S.; Franklin, M.J.; Papke, R.T. Biofilms formed by the archaeon Haloferax volcaniiexhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 2014, 12, 65. [Google Scholar] [CrossRef]
- Dueholm, M.S.; Larsen, P.; Finster, K.; Stenvang, M.R.; Christiansen, G.; Vad, B.S.; Bøggild, A.; Otzen, D.E.; Nielsen, P.H. The Tubular Sheaths Encasing Methanosaeta thermophila Filaments Are Functional Amyloids. J. Biol. Chem. 2015, 290, 20590–20600. [Google Scholar] [CrossRef]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia coli Curli Operons in Directing Amyloid Fiber Formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Søndergaard, M.T.; Nilsson, M.; Christiansen, G.; Stensballe, A.; Overgaard, M.T.; Givskov, M.; Tolker-Nielsen, T.; Otzen, D.E.; Nielsen, P.H. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2013, 2, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.; Nielsen, J.L.; Dueholm, M.S.; Wetzel, R.; Otzen, D.; Nielsen, P.H. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007, 9, 3077–3090. [Google Scholar] [CrossRef]
- Elliot, M.A.; Karoonuthaisiri, N.; Huang, J.; Bibb, M.J.; Cohen, S.N.; Kao, C.M.; Buttner, M.J. The chaplins: A family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003, 17, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Willemse, J.; Sawyer, E.B.; Lou, F.; Gong, W.; Zhang, H.; Gras, S.L.; Claessen, D.; Perrett, S. The propensity of the bacterial rodlin protein RdlB to form amyloid fibrils determines its function in Streptomyces coelicolor. Sci. Rep. 2017, 7, 42867. [Google Scholar] [CrossRef]
- Bieler, S.; Estrada, L.; Lagos, R.; Baeza, M.; Castilla, J.; Soto, C. Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 2005, 280, 26880–26885. [Google Scholar] [CrossRef]
- Bavdek, A.; Kostanjšek, R.; Antonini, V.; Lakey, J.H.; Dalla Serra, M.; Gilbert, R.J.C.; Anderluh, G. pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 2012, 279, 126–141. [Google Scholar] [CrossRef]
- Oh, J.; Kim, J.-G.; Jeon, E.; Yoo, C.-H.; Moon, J.S.; Rhee, S.; Hwang, I. Amyloidogenesis of type III-dependent harpins from plant pathogenic bacteria. J. Biol. Chem. 2007, 282, 13601–13609. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Spaink, H.P.; Kondorosi, A.; Hooykaas, P. The Rhizobiaceae: Molecular Biology of Model Plant-Associated Bacteria; Springer: Dordrecht, The Netherlands, 1998; ISBN 978-0-7923-5180-1. [Google Scholar] [CrossRef]
- Hentschel, U.; Steinert, M.; Hacker, J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000, 8, 226–231. [Google Scholar] [CrossRef]
- Deakin, W.J.; Broughton, W.J. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009, 7, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Alexandrov, A.I.; Ryzhova, T.A.; Mitkevich, O.V.; Dergalev, A.A.; Ter-Avanesyan, M.D.; Galkin, A.P. Proteomic screening for amyloid proteins. PLoS ONE 2014, 9, e116003. [Google Scholar] [CrossRef] [PubMed]
- Afonin, A.; Sulima, A.; Zhernakov, A.I.; Zhukov, V.A. Draft genome of the strain RCAM1026 Rhizobium leguminosarum bv. viciae. Genom. Data 2016, 11, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Nizhnikov, A.A.; Ryzhova, T.A.; Volkov, K.V.; Zadorsky, S.P.; Sopova, J.V.; Inge-Vechtomov, S.G.; Galkin, A.P. Interaction of Prions Causes Heritable Traits in Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006504. [Google Scholar] [CrossRef]
- Antonets, K.S.; Volkov, K.V.; Maltseva, A.L.; Arshakian, L.M.; Galkin, A.P.; Nizhnikov, A.A. Proteomic Analysis of Escherichia coli Protein Fractions Resistant to Solubilization by Ionic Detergents. Biochemistry (Moscow) 2016, 81, 34–46. [Google Scholar] [CrossRef]
- Sivanathan, V.; Hochschild, A. A bacterial export system for generating extracellular amyloid aggregates. Nat. Protoc. 2013, 8, 1381–1390. [Google Scholar] [CrossRef]
- Sivanathan, V.; Hochschild, A. Generating extracellular amyloid aggregates using E. coli cells. Genes Dev. 2012, 26, 2659–2667. [Google Scholar] [CrossRef]
- Antonets, K.S.; Belousov, M.V.; Belousova, M.E.; Nizhnikov, A.A. The Gln3 Transcriptional Regulator of Saccharomyces cerevisiae Manifests Prion-Like Properties upon Overproduction. Biochemistry (Moscow) 2019, 84, 441–451. [Google Scholar] [CrossRef]
- Antonets, K.S.; Sargsyan, H.M.; Nizhnikov, A.A. A Glutamine/Asparagine-Rich Fragment of Gln3, but not the Full-Length Protein, Aggregates in Saccharomyces cerevisiae. Biochemistry (Moscow) 2016, 81, 407–413. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Belousov, M.V.; Bondarev, S.A.; Moskalenko, S.E.; Zhouravleva, G.A. Identification of new genes that affect [PSI+] prion toxicity in Saccharomyces cerevisiae yeast. Mol. Biol. (Mosc.) 2016, 50, 710–718. [Google Scholar] [CrossRef]
- Kaiser, C.; Michaelis, S.; Mitchell, A.; Cold Spring Harbor Laboratory. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1994; ISBN 978-0879694517. [Google Scholar]
- Serio, T.R.; Cashikar, A.G.; Moslehi, J.J.; Kowal, A.S.; Lindquist, S.L. [41] Yeast prion [Ψ+] and its determinant, sup35p. In Methods in Enzymology; Elsevier: New York, NY, USA, 1999; Volume 309, pp. 649–673. [Google Scholar] [CrossRef]
- Kayed, R.; Head, E.; Sarsoza, F.; Saing, T.; Cotman, C.W.; Necula, M.; Margol, L.; Wu, J.; Breydo, L.; Thompson, J.L.; et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, I.M.; Sulatskaya, A.I.; Uversky, V.N.; Turoverov, K.K. A new trend in the experimental methodology for the analysis of the thioflavin T binding to amyloid fibrils. Mol. Neurobiol. 2012, 45, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Sulatskaya, A.I.; Rodina, N.P.; Povarova, O.I.; Kuznetsova, I.M.; Turoverov, K.K. Different conditions of fibrillogenesis cause polymorphism of lysozyme amyloid fibrils. J. Mol. Struct. 2017, 1140, 52–58. [Google Scholar] [CrossRef]
- Levine, M.M.; Ristaino, P.; Marley, G.; Smyth, C.; Knutton, S.; Boedeker, E.; Black, R.; Young, C.; Clements, M.L.; Cheney, C. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: Morphology, purification, and immune responses in humans. Infect. Immun. 1984, 44, 409–420. [Google Scholar]
- Sacher, J. Immunogold labeling of bacterial cells for transmission electron microscopy (TEM). protocols.io 2018. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Habour: New York, NY, USA, 1989; ISBN 0879693096. [Google Scholar]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and analysis of prion and amyloid aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef]
- Halfmann, R.; Lindquist, S. Screening for amyloid aggregation by semi-denaturing detergent-agarose gel electrophoresis. J. Vis. Exp. 2008, 17, 838. [Google Scholar] [CrossRef]
- Vladimirov, Y.; Litvin, F.F. Photobiology and Spectroscopic Methods; Springer: Berlin, Germany, 1964. [Google Scholar]
- Fonin, A.V.; Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS ONE 2014, 9, e103878. [Google Scholar] [CrossRef]
- O’Connor, D.V.P. Time-Correlated Single Photon Counting; Academic Press: New York, NY, USA, 1984; pp. 37–54. [Google Scholar]
- Marquardt, D.W. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. J. Soc. Ind. Appl. Math. 2005, 11, 431–441. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Goers, J.; Permyakov, S.E.; Permyakov, E.A.; Uversky, V.N.; Fink, A.L. Conformational prerequisites for α-lactalbumin fibrillation. Biochemistry 2002, 41, 12546–12551. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, I.; Bustamante, C.; Maestre, M.F. The optical activity of nucleic acids and their aggregates. Annu. Rev. Biophys. Bioeng. 1980, 9, 107–141. [Google Scholar] [CrossRef]
- LeVine, H., 3rd. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999, 309, 274–284. [Google Scholar] [CrossRef]
- LeVine, H., 3rd. Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Maskevich, A.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Fluorescence quantum yield of thioflavin T in rigid isotropic solution and incorporated into the amyloid fibrils. PLoS ONE 2010, 5, e15385. [Google Scholar] [CrossRef]
- Krebs, M.R.H.; Bromley, E.H.C.; Donald, A.M. The binding of thioflavin-T to amyloid fibrils: Localisation and implications. J. Struct. Biol. 2005, 149, 30–37. [Google Scholar] [CrossRef]
- Sulatskaya, A.I.; Lavysh, A.V.; Maskevich, A.A.; Kuznetsova, I.M.; Turoverov, K.K. Thioflavin T fluoresces as excimer in highly concentrated aqueous solutions and as monomer being incorporated in amyloid fibrils. Sci. Rep. 2017, 7, 2146. [Google Scholar] [CrossRef]
- Zaat, S.A.J.; Wijffelman, C.A.; Spaink, H.P.; van Brussel, A.A.; Okker, R.J.; Lugtenberg, B.J. Induction of the nodA promoter of Rhizobium leguminosarum sym plasmid pRL1JI by plant flavanones and flavones. J. Bacteriol. 1987, 169, 198–204. [Google Scholar] [CrossRef]
- Tolin, S.; Arrigoni, G.; Moscatiello, R.; Masi, A.; Navazio, L.; Sablok, G.; Squartini, A. Quantitative analysis of the naringenin-inducible proteome in Rhizobium leguminosarum by isobaric tagging and mass spectrometry. Proteomics 2013, 13, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Dudman, W.F. Capsulation in Rhizobium species. J. Bacteriol. 1968, 95, 1200–1201. [Google Scholar] [PubMed]
- Wösten, H.A.B.; de Vocht, M.L. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 2000, 1469, 79–86. [Google Scholar] [CrossRef]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef]
- Van Gerven, N.; Van der Verren, S.E.; Reiter, D.M.; Remaut, H. The Role of Functional Amyloids in Bacterial Virulence. J. Mol. Biol. 2018, 430, 3657–3684. [Google Scholar] [CrossRef]
- Taglialegna, A.; Navarro, S.; Ventura, S.; Garnett, J.A.; Matthews, S.; Penades, J.R.; Lasa, I.; Valle, J. Staphylococcal Bap Proteins Build Amyloid Scaffold Biofilm Matrices in Response to Environmental Signals. PLoS Pathog. 2016, 12, e1005711. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Yuan, A.H.; Hochschild, A. A bacterial global regulator forms a prion. Science 2017, 355, 198–201. [Google Scholar] [CrossRef]
- Kaur, G.; Kaundal, S.; Kapoor, S.; Grimes, J.M.; Huiskonen, J.T.; Thakur, K.G. Mycobacterium tuberculosis CarD, an essential global transcriptional regulator forms amyloid-like fibrils. Sci. Rep. 2018, 8, 10124. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Kapoor, S.; Thakur, K.G. Bacillus subtilis HelD, an RNA Polymerase Interacting Helicase, Forms Amyloid-Like Fibrils. Front. Microbiol. 2018, 9, 1934. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Son, M.; Edskes, H.K. Prion variants of yeast are numerous, mutable, and segregate on growth, affecting prion pathogenesis, transmission barriers, and sensitivity to anti-prion systems. Viruses 2019, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Vlamakis, H.; Losick, R.; Koltera, R. Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J. Bacteriol. 2014, 196, 1505–1513. [Google Scholar] [CrossRef]
- Romero, D.; Vlamakis, H.; Losick, R.; Kolter, R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol. Microbiol. 2011, 80, 1155–1168. [Google Scholar] [CrossRef]
- Erskine, E.; Morris, R.J.; Schor, M.; Earl, C.; Gillespie, R.M.C.; Bromley, K.M.; Sukhodub, T.; Clark, L.; Fyfe, P.K.; Serpell, L.C.; et al. Formation of functional, non-amyloidogenic fibres by recombinant Bacillus subtilis TasA. Mol. Microbiol. 2018, 110, 897–913. [Google Scholar] [CrossRef]
- Galdiero, S.; Falanga, A.; Cantisani, M.; Tarallo, R.; Elena Della Pepa, M.; D’Oriano, V.; Galdiero, M. Microbe-Host Interactions: Structure and Role of Gram-Negative Bacterial Porins. Curr. Protein Pept. Sci. 2012, 13, 843–854. [Google Scholar] [CrossRef]
- De Maagd, R.A.; Yang, W.C.; Goosen-de Roo, L.; Mulders, I.H.M.; Roest, H.P.; Spaink, H.P.; Bisseling, T.; Lugtenberg, B.J.J. Down-regulation of expression of the Rhizobium leguminosarum outer membrane protein gene ropA occurs abruptly in interzone II-III of pea nodules and can be uncoupled from nif gene activation. Mol. Plant-Microbe Interact. 1994, 7, 276–281. [Google Scholar]
- Roest, H.P.; Mulders, I.H.; Wijffelman, C.A.; Lugtenberg, B.J. Isolation of ropB, a Gene Encoding a 22-kDa Rhizobium leguminosarum Outer Membrane Protein. Mol. Plant-Microbe Interact. 1995, 8, 576–583. [Google Scholar]
- Fraysse, N.; Couderc, F.; Poinsot, V. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur. J. Biochem. 2003, 270, 1365–1380. [Google Scholar] [CrossRef]
- Ryzhova, T.A.; Sopova, J.V.; Zadorsky, S.P.; Siniukova, V.A.; Sergeeva, A.V.; Galkina, S.A.; Nizhnikov, A.A.; Shenfeld, A.A.; Volkov, K.V.; Galkin, A.P. Screening for amyloid proteins in the yeast proteome. Curr. Genet. 2018, 64, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Danoff, E.J.; Fleming, K.G. Aqueous, Unfolded OmpA forms amyloid-like fibrils upon self-association. PLoS ONE 2015, 10, e0132301. [Google Scholar] [CrossRef] [PubMed]
- Joseph Sahaya Rajan, J.; Chinnappan Santiago, T.; Singaravel, R.; Ignacimuthu, S. Outer membrane protein C (OmpC) of Escherichia coli induces neurodegeneration in mice by acting as an amyloid. Biotechnol. Lett. 2016, 38, 689–700. [Google Scholar] [CrossRef]
- Meuskens, I.; Michalik, M.; Chauhan, N.; Linke, D.; Leo, J.C. A new strain collection for improved expression of outer membrane proteins. Front. Cell. Infect. Microbiol. 2017, 7, 464. [Google Scholar] [CrossRef]
- Montes García, J.F.; Vaca, S.; Delgado, N.L.; Uribe-García, A.; Vázquez, C.; Sánchez Alonso, P.; Xicohtencatl Cortes, J.; Cruz Cordoba, A.; Negrete Abascal, E. Mannheimia haemolytica OmpP2-like is an amyloid-like protein, forms filaments, takes part in cell adhesion and is part of biofilms. Antonie van Leeuwenhoek 2018, 111, 2311–2321. [Google Scholar] [CrossRef]
- Villain, E.; Nikekhin, A.A.; Kajava, A.V. Porins and Amyloids are Coded by Similar Sequence Motifs. Proteomics 2019, 19, e1800075. [Google Scholar] [CrossRef]
- Antonets, K.S.; Nizhnikov, A.A. Predicting amyloidogenic proteins in the proteomes of plants. Int. J. Mol. Sci. 2017, 18, 2155. [Google Scholar] [CrossRef]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef]
- Rivilla, R.; Sutton, J.M.; Downie, J.A. Rhizobium leguminosarum NodT is related to a family of outer-membrane transport proteins that includes TolC, PrtF, CyaE and AprF. Gene 1995, 161, 27–31. [Google Scholar] [CrossRef]
- Antonets, K.S.; Kliver, S.F.; Nizhnikov, A.A. Exploring Proteins Containing Amyloidogenic Regions in the Proteomes of Bacteria of the Order Rhizobiales. Evol. Bioinform. 2018, 14. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosolapova, A.O.; Belousov, M.V.; Sulatskaya, A.I.; Belousova, M.E.; Sulatsky, M.I.; Antonets, K.S.; Volkov, K.V.; Lykholay, A.N.; Shtark, O.Y.; Vasileva, E.N.; et al. Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum. Biomolecules 2019, 9, 694. https://doi.org/10.3390/biom9110694
Kosolapova AO, Belousov MV, Sulatskaya AI, Belousova ME, Sulatsky MI, Antonets KS, Volkov KV, Lykholay AN, Shtark OY, Vasileva EN, et al. Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum. Biomolecules. 2019; 9(11):694. https://doi.org/10.3390/biom9110694
Chicago/Turabian StyleKosolapova, Anastasiia O., Mikhail V. Belousov, Anna I. Sulatskaya, Maria E. Belousova, Maksim I. Sulatsky, Kirill S. Antonets, Kirill V. Volkov, Anna N. Lykholay, Oksana Y. Shtark, Ekaterina N. Vasileva, and et al. 2019. "Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum" Biomolecules 9, no. 11: 694. https://doi.org/10.3390/biom9110694
APA StyleKosolapova, A. O., Belousov, M. V., Sulatskaya, A. I., Belousova, M. E., Sulatsky, M. I., Antonets, K. S., Volkov, K. V., Lykholay, A. N., Shtark, O. Y., Vasileva, E. N., Zhukov, V. A., Ivanova, A. N., Zykin, P. A., Kuznetsova, I. M., Turoverov, K. K., Tikhonovich, I. A., & Nizhnikov, A. A. (2019). Two Novel Amyloid Proteins, RopA and RopB, from the Root Nodule Bacterium Rhizobium leguminosarum. Biomolecules, 9(11), 694. https://doi.org/10.3390/biom9110694










