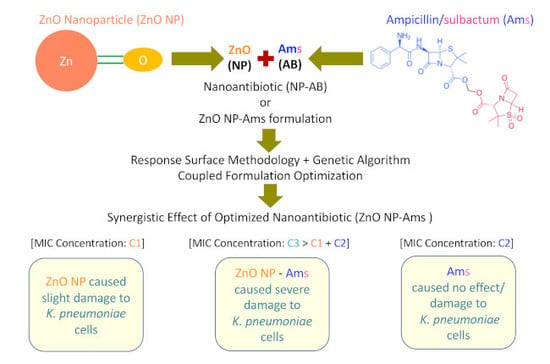
Preparation and Evaluation of the ZnO NP–Ampicillin/Sulbactam Nanoantibiotic: Optimization of Formulation Variables Using RSM Coupled GA Method and Antibacterial Activities
Abstract
1. Introduction
2. Results
2.1. Selection of Drug for Nanoantibiotic Formulation
2.2. Minimum Inhibitory Concentration (MIC) of Ams against Resistant Bacterial Strains
2.3. Antibacterial Activity of ZnO NP
2.4. ‘ZnO NP–Ams’ Nanoantibiotics—Formulation and Optimization Employing Statistical Design
2.4.1. ANOVA Analysis
2.4.2. Contour Plots
2.5. Genetic Algorithm-Based Optimization
2.6. ROS Estimation
2.7. Determination of MIC of Optimized ZnO NP-Ampicillin/Sulbactam Nanoantibiotic
2.8. Scanning Electron Microscopy
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains, Culture Conditions and Antibiotics
5.2. Antibiotic Resistance Profile of Bacterial Strains
5.3. Minimum Inhibitory Concentration of Antibiotic Against Different Bacterial Strains
5.4. Activity of ZnO NP against Different Bacterial Strains
5.5. Formulation and Optimization of Nanoantibiotics
5.6. GA Optimization
5.7. Estimation of Reactive Oxygen Species
5.8. Scanning Electron Microscopic Examinations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wise, R.; Hart, T.; Cars, O.; Streulens, M.; Helmuth, R.; Huovinen, P.; Sprenger, M. Antimicrobial resistance: is a major threat to public health. Br. Med. J. 1998, 317, 609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gu, F.X.; Chan, J.M.; Wang, A.Z.; Langer, R.S.; Farokhzad, O.C. Nanoparticles in medicine: therapeutic applications and developments. Clin. Pharm. 2008, 83, 761–769. [Google Scholar]
- Hong, B.; Kai, J.; Ren, Y.; Zou, Z.; Ahn, C.H.; Kang, K.A. Highly sensitive rapid, reliable, and automatic cardiovascular disease diagnosis with nanoparticle fluorescence enhancer and mems. In Oxygen Transport to Tissue XXIX; Springer: Boston, MA, USA, 2008; pp. 265–273. [Google Scholar]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Van Giau, V.; Ann, S.S.; Hulme, J. Recent advances in the treatment of pathogenic infections using antibiotics and nano-drug delivery vehicles. Drug Design Dev. Ther. 2019, 13, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Curtis, A.; Hoskins, C. Application of nanoparticle technologies in the combat against anti-microbial resistance. Pharmaceutics 2018, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyev, A.M.; Kon, K.V.; Abamor, E.S.; Bagirova, M.; Rafailovich, M. Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev. Anti Infect. 2011, 9, 1035–1052. [Google Scholar]
- Shahverdi, A.R.; Fakhimi, A.; Shahverdi, H.R.; Minaian, S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 168–171. [Google Scholar] [CrossRef]
- Sawai, J. Quantitative evaluation of antibacterial activities of metallic oxide powders (ZnO, MgO and CaO) by conductimetric assay. J. Microbiol. Methods 2003, 54, 177–182. [Google Scholar] [CrossRef]
- Sawai, J.; Yoshikawa, T. Quantitative evaluation of antifungal activity of metallic oxide powders (MgO, CaO and ZnO) by an indirect conductimetric assay. J. Appl. Microbiol. 2004, 96, 803–809. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, Y.; Povey, M.; York, D. ZnO nanofluids—A potential antibacterial agent. Prog. Nat. Sci. 2008, 18. [Google Scholar] [CrossRef]
- Jones, N.; Ray, B.; Ranjit, K.T.; Manna, A.C. Antibacterial activity of ZnO nanoparticle suspensions on a broad spectrum of microorganisms. Fems Microbiol. Lett. 2008, 279, 71–76. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, T.; Wei, D.; Wei, Y.; Li, Y.; Zhang, H. Core–shell nanocarriers with ZnO quantum dots-conjugated Au nanoparticle for tumor-targeted drug delivery. Carbohydr. Polym. 2013, 92, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Lansdown, A.B.; Mirastschijski, U.; Stubbs, N.; Scanlon, E.; Ågren, M.S. Zinc in wound healing: theoretical, experimental, and clinical aspects. Wound Repair Regen. 2007, 15, 2–16. [Google Scholar] [PubMed]
- FDA (Food and Drug Administration). Select Committee on GRAS Substances (SCOGS) Opinion: Zinc Salts 2015; Food and Drug Administration: Washington DC, USA, 2010. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=182.8991 (accessed on 5 November 2018).
- Singh, V.; Haque, S.; Niwas, R.; Srivastava, A.; Pasupuleti, M.; Tripathi, C.K.M. Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 2017, 7, 2087. [Google Scholar]
- Srivastava, A.; Singh, V.; Pandey, S.; Mishra, M.; Jawed, A.; Shukla, P.K.; Singh, P.K.; Tripathi, C.K.M. Response surface methodology-Genetic algorithm based medium optimization, purification, and characterization of Cholesterol Oxidase from Streptomyces Rimosus. Sci. Rep. 2018, 8, 10913. [Google Scholar] [CrossRef] [PubMed]
- Dubey, K.K.; Jawed, A.; Haque, S. Enhanced extraction of 3- demethylated colchicine from fermentation broth of Bacillus megaterium: optimization of process parameters by statistical experimental design. Eng. Life Sci. 2011, 11, 598–606. [Google Scholar] [CrossRef]
- Haque, S.; Khan, S.; Wahid, M.; Dar, S.A.; Soni, S.; Mandal, R.K.; Singh, V.; Tiwari, D.; Lohani, M.; Areeshi, M.Y.; et al. Artificial intelligence vs. Statistical modeling and optimization of continuous bead milling process for bacterial cell lysis. Front. Microbiol. 2016, 7, 1852. [Google Scholar] [CrossRef] [PubMed]
- Arafa, M.G.; Ayoub, B.M. DOE Optimization of nano-based carrier of Pregabalin as hydrogel: New therapeutic & chemometric approaches for controlled drug delivery systems. Sci. Rep. 2017, 7, 41503. [Google Scholar] [CrossRef] [PubMed]
- Ojha, S.; Kumar, B. Formulation and optimization of chitosan nanoparticles of dimethyl fumarate using Box-Behnken Design. Int. J. App. Pharm. 2016, 8, 10–17. [Google Scholar]
- Wu, Z.; Guan, R.; Lyu, F.; Liu, M.; Gao, J.; Cao, G. Optimization of preparation conditions for lysozyme nanoliposomes using response surface methodology and evaluation of their stability. Molecules 2016, 21, 741. [Google Scholar] [CrossRef]
- Honary, S.; Ebrahimi, P.; Hadianamrei, R. Optimization of particle size and encapsulation efficiency of vancomycin nanoparticles by Response Surface methodology. Pharm. Dev. Technol. 2014, 19, 987–998. [Google Scholar] [CrossRef]
- Liu, S.; Ho, P.C. Formulation optimization of scutellarin-loaded HP-β-CD/chitosan nanoparticles using response surface methodology with Box–Behnken design. Asian J. Pharm. Sci. 2017, 12, 378–385. [Google Scholar] [CrossRef]
- Boonyasirisri, P.; Nimmannit, U.; Rojsitthisak, P.; Bhunchu, S.; Rojsitthisak, P. Optimization of curcuminoid-loaded PLGA nanoparticles using Box-Behnken statistical design. J. Nano Res. 2015, 33, 60–71. [Google Scholar] [CrossRef]
- Carraro, T.C.; Khalil, N.M.; Mainardes, R.M. Amphotericin B-loaded polymeric nanoparticles: Formulation optimization by factorial design. Pharm. Dev. Technol. 2016, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Petri, W.A.; Brunton, L.L.; Chabner, B.A.; Knollmann, B.C. Goodman and Gilman’s The Pharmacological Basis of Therapeutics, 12th ed.; McGraw-Hill: New York, NY, USA, 1996; Chapter 53. [Google Scholar]
- Drawz, S.M.; Bonomo, R.A. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef] [PubMed]
- Adnan, S.; Paterson, D.L.; Lipman, J.; Roberts, J.A. Ampicillin/sulbactam: its potential use in treating infections in critically ill patients. Int. J. Antimicrob. Agents 2013, 42, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Rupp, M.E.; Fey, P.D. Extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Drugs 2003, 63, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Jabalameli, F.; Mirsalehian, A.; Sotoudeh, N.; Jabalameli, L.; Aligholi, M.; Khoramian, B.; Taherikalani, M.; Emaneini, M. Multiple-locus variable number of tandem repeats 86 (VNTR) fingerprinting (MLVF) and antibacterial resistance profiles of extended spectrum beta lactamase (ESBL) producing Pseudomonas aeruginosa among burnt patients in Tehran. Burns 2011, 37, 1202‘–1207. [Google Scholar] [CrossRef]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Med. 2006, 119, S20–S28. [Google Scholar] [CrossRef]
- Kruger, T.; Szabo, D.; Keddy, K.H.; Deeley, K.; Marsh, J.W.; Hujer, A.M.; Bonomo, R.A.; Paterson, D.L. Infections with nontyphoidal Salmonella species producing TEM-63 or a novel TEM enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 2004, 48, 4263–4270. [Google Scholar] [CrossRef]
- Rodríguez-Baño, J.; Navarro, M.D.; Romero, L.; Martínez-Martínez, L.; Muniain, M.A.; Perea, E.J.; Pérez-Cano, R.; Pascual, A. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J. Clin. Microbiol. 2004, 42, 1089–1094. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab J. Chem. 2017. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.; Ann, L.C.; Bakhori, S.K.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef]
- Pathak, L.; Singh, V.; Niwas, R.; Osama, K.; Khan, S.; Haque, S.; Tripathi, C.K.M.; Mishra, B.N. Artificial intelligence versus statistical modeling and optimization of cholesterol oxidase production by using Streptomyces sp. Plos ONE 2015, 10, e0137268. [Google Scholar] [CrossRef] [PubMed]
- Hussein-Al-Ali, S.H.; El Zowalaty, M.E.; Hussein, M.Z.; Geilich, B.M.; Webster, T.J. Synthesis, characterization, and antimicrobial activity of an ampicillin-conjugated magnetic nanoantibiotic for medical applications. Int. J. Nanomed. 2014, 9, 3801–3814. [Google Scholar] [CrossRef] [PubMed]
- Abed, N.; Saïd-Hassane, F.; Zouhiri, F.; Mougin, J.; Nicolas, V.; Desmaële, D.; Gref, R.; Couvreur, P. An efficient system for intracellular delivery of beta-lactam antibiotics to overcome bacterial resistance. Sci. Rep. 2015, 5, 13500. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zheng, X.; Yan, D.; Yin, G.; Liao, X.; Kang, Y.; Yao, Y.; Huang, D.; Hao, B. Toxicological effect of ZnO nanoparticles based on bacteria. Langmuir 2008, 24, 4140–4144. [Google Scholar] [CrossRef]
- Wang, L.; Hu., C.; Shao, L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Arakha, M.; Saleem, M.; Mallickkha, B.C.; Jha, S. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci. Rep. 2015, 5, 9578. [Google Scholar] [CrossRef]
- Aranda, A.; Sequedo, L.; Tolosa, L.; Quintas, G.; Burello, E.; Castell, J.V.; Gombau, L. Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay: A quantitative method for oxidative stress assessment of nanoparticle-treated cells. Toxicol Vitr. 2013, 27, 954–963. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Singh, S.P.; Häder, D.P.; Sinha, R.P. Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2’,7’-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem. Biophys. Res. Commun. 2010, 397, 603–607. [Google Scholar] [CrossRef]
- Tiwari, V.; Mishra, N.; Gadani, K.; Solanki, P.S.; Shah, N.; Tiwari, M. Mechanism of anti-bacterial activity of Zinc Oxide nanoparticle against carbapenem resistant Acinetobacter baumannii. Front. Microbiol. 2018, 9, 1218. [Google Scholar] [CrossRef]
- Yi, G.; Li, X.; Yuan, Y.; Zhang, Y. Redox active Zn/ZnO duo generating superoxide ( O2−) and H2O2 under all conditions for environmental sanitation. Environ. Sci. Nano 2019, 6, 68–74. [Google Scholar] [CrossRef]
- Punnoose, A.; Dodge, K.; Rasmussen, J.W.; Chess, J.; Wingett, D.; Anders, C. Cytotoxicity of ZnO nanoparticles can be tailored by modifying their surface structure: A green chemistry approach for safer nanomaterials. ACS Sustain. Chem. Eng. 2014, 2, 1666–1673. [Google Scholar] [CrossRef]
- Li, M.; Zhu, L.; Lin, D. Toxicity of ZnO nanoparticles to Escherichia coli: Mechanism and the influence of medium components. Env. Sci. Technol. 2011, 45, 1977–1983. [Google Scholar] [CrossRef]
- Lipovsky, A.; Nitzan, Y.; Gedanken, A.; Lubart, R. Antifungal activity of ZnO nanoparticles—The role of ROS mediated cell injury. Nanotechnology 2011, 22, 105101. [Google Scholar] [CrossRef] [PubMed]
- Siwinska-Stefanska, K.; Kubiaka, A.; Piasecki, A.; Goscianska, J.; Nowaczyk, G.; Jurga, S.; Jesionowski, T. TiO2-ZnO binary oxide systems: Comprehensive characterization and tests of photocatalytic activity. Materials 2018, 11, 841. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Jia, H.; Cai, J.; Han, X.; Zheng, Z.; Wamer, W.G.; Yin, J.-J. Production of reactive oxygen species and electrons from photoexcited ZnO and ZnS nanoparticles: A comparative study for unraveling their distinct photocatalytic activities. J. Phys. Chem. C. 2016, 120, 3187–3195. [Google Scholar] [CrossRef]
- Sivakumar, P.; Lee, M.; Kim, Y.-S.; Shim, M.S. Photo-triggered antibacterial and anticancer activities of zinc oxide nanoparticles. J. Mater. Chem. B. 2018, 6, 4852–4871. [Google Scholar] [CrossRef]
- Banoee, M.; Seif, S.; Nazari, Z.E.; Jafari-Fesharaki, P.; Shahverdi, H.R.; Moballegh, A.; Moghaddam, K.M.; Shahverdi, A.R. ZnO nanoparticles enhanced antibacterial activity of ciprofloxacin against staphylococcus aureus and Escherichia coli. J. Biomed. Mater. Res. B Appl. Biomater. 2010, 93, 557–561. [Google Scholar] [CrossRef]
- Navale, G.R.; Thripuranthaka, M.; Late, D.J.; Shinde, S.S. Antimicrobial activity of ZnO nanoparticles against pathogenic bacteria and fungi. JSM Nanotechnol. Nanomed. 2015, 3, 1033. [Google Scholar]
- Khan, I.; Bahuguna, A.; Kumar, P.; Bajpai, V.K.; Kang, S.C. Antimicrobial potential of carvacrol against uropathogenic Escherichia coli via membrane disruption, depolarization, and reactive oxygen species generation. Front. Microbiol. 2017, 8, 2421. [Google Scholar] [CrossRef]
- Hameed, A.S.; Karthikeyan, C.; Ahamed, A.P.; Thajuddin, N.; Alharbi, N.S.; Alharbi, S.A.; Ravi, G. In vitro antibacterial activity of ZnO and Nd doped ZnO nanoparticles against ESBL producing Escherichia coli and Klebsiella pneumoniae. Sci. Rep. 2016, 6, 24312. [Google Scholar] [CrossRef]
- Edoo, Z.; Arthur, M.; Hugonnet, J.E. Reversible inactivation of a peptidoglycan transpeptidase by a β-lactam antibiotic mediated by β-lactam-ring recyclization in the enzyme active site. Sci. Rep. 2017, 7, 9136. [Google Scholar] [CrossRef]
- Pati, R.; Mehta, R.K.; Mohanty, S.; Padhi, A.; Sengupta, M.; Vaseeharan, B.; Goswami, C.; Sonawane, A. Topical application of zinc oxide nanoparticles reduces bacterial skin infection in mice and exhibits antibacterial activity by inducing oxidative stress response and cell membrane disintegration in macrophages. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1195–1208. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Wikler, M.A. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 5th ed.National Committee for Clinical Laboratory Standards (NCCLS): Wayne, PA, USA, 2000. [Google Scholar]
- Moballegh, A.; Shahverdi, H.R.; Aghababazadeh, R.; Mirhabibi, A.R. ZnO nanoparticles obtained by mechanochemical technique and the optical properties. Surf. Sci 2007, 601, 2850–2854. [Google Scholar] [CrossRef]
- Prasad, R.G.S.V.; Basavaraju, D.; Rao, K.N.; Naveen, C.S.; Endrino, J.; Phani, A.R. Nanostructured TiO2 and TiO2-Ag antimicrobial thin films. In Proceedings of 2011 International Conference on Nanoscience, Technology and Societal Implications (NSTSI), Bhubaneswar, India, 8–10 December 2011; pp. 1–6. [Google Scholar] [CrossRef]




| S. No. | Antibiotic (Concentration) | Zone of Inhibition (mm) | |||||
|---|---|---|---|---|---|---|---|
| S. a | E. c | B. p | S. t | K. p | P. a | ||
| 1. | Amikacin (Ak30) | 30 | 22 | 25 | 26 | 29 | 19 |
| 2. | Ampicillin (A10) | R | 10 | 19 | R | R | R |
| 3. | Ampicillin/Sulbactam (As10) | R | R | 11 | R | R | R |
| 4. | Amoxyclav (Ac30) | R | 21 | 24 | R | R | R |
| 5. | Ceftazidime (Ca30) | 30 | R | R | R | 13 | R |
| 6. | Cephotaxime (Ce30) | R | R | 18 | 11 | R | 11 |
| 7. | Ciprofloxacin (Cf5) | 19 | 30 | 24 | 25 | 31 | 29 |
| 8. | Clindamycin (Cd2) | 35 | R | 21 | 20 | 33 | 10 |
| 9. | Co-Trimoxazole (Co25) | R | 23 | 33 | 24 | R | 29 |
| 10. | Erythromycin (E15) | 28 | 10 | R | 10 | 31 | 20 |
| 11. | Gentamycin (G10) | 26 | 23 | 22 | 18 | 24 | 11 |
| 12. | Nalidixic acid (Na30) | 11 | R | 26 | 18 | 25 | 30 |
| 13. | Netillin (Nt30) | 12 | 13 | 15 | 12 | 17 | 11 |
| 14. | Nitrofurantoin (Nf300) | R | 20 | 23 | 21 | 18 | 10 |
| 15. | Penicillin G (P10) | R | 20 | 26 | R | R | R |
| 16. | Tobramycin (Tb10) | 15 | 13 | 20 | 15 | 20 | 11 |
| 17. | Vancomycin (Va30) | R | 23 | 21 | 16 | 19 | 16 |
| S. No. | Bacterial Strains | MIC (μg/mL) |
|---|---|---|
| 1. | Escherichia coli MTCC 1304 | 50 |
| 2. | Klebsiella pneumoniae MTCC 3384 | 100 |
| 3. | Pseudomonas aeruginosa MTCC 741 | 100 |
| 4. | Salmonella typhi MTCC 537 | 50 |
| 5. | Staphylococcus aureus MTCC 902 | 50 |
| S. No. | Bacterial Strains | Zone of Inhibition (in mm) at Different Concentration (µg) | |||
|---|---|---|---|---|---|
| 25 | 50 | 100 | 200 | ||
| 1. | Escherichia coli MTCC 1304 | 9 | 10 | 11 | 13 |
| 2. | Klebsiella pneumoniae MTCC 3384 | 19 | 20 | 22 | 25 |
| 3. | Pseudomonas aeruginosa MTCC 741 | 5 | 6 | 8 | 10 |
| 4. | Salmonella typhi MTCC 537 | 12 | 14 | 18 | 20 |
| 5. | Staphylococcus aureus MTCC 902 | 7 | 10 | 14 | 16 |
| Runs | X1 Coded Uncoded | X1 Coded Uncoded | X1 Coded Uncoded | ZOI (mm) Experimental Predicted Residual | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1. | +1 | 80 | +1 | 65 | +1 | 36 | 26 | 24.56 | 1.44 |
| 2. | +1 | 80 | −1 | 25 | −1 | 12 | 25 | 23.06 | 1.94 |
| 3. | −1 | 30 | +1 | 65 | −1 | 12 | 22 | 21.81 | 0.19 |
| 4. | +1 | 80 | +1 | 65 | −1 | 12 | 27 | 26.81 | 0.19 |
| 5. | +1 | 80 | −1 | 25 | +1 | 36 | 25 | 23.81 | 1.19 |
| 6. | −1 | 30 | +1 | 65 | +1 | 36 | 20 | 20.56 | 0.56 |
| 7. | −1 | 30 | −1 | 25 | +1 | 36 | 25 | 23.81 | 1.19 |
| 8. | −1 | 30 | −1 | 25 | −1 | 12 | 22 | 22.06 | 0.06 |
| 9. | −2 | 5 | 0 | 45 | 0 | 24 | 21 | 20.68 | 0.32 |
| 10. | 0 | 55 | −2 | 5 | 0 | 24 | 21 | 22.43 | 1.43 |
| 11. | 0 | 55 | 0 | 45 | −2 | 0 | 21 | 21.43 | 0.43 |
| 12. | +2 | 105 | 0 | 45 | 0 | 24 | 24 | 25.68 | 1.68 |
| 13. | 0 | 55 | +2 | 85 | 0 | 24 | 23 | 22.93 | 0.07 |
| 14. | 0 | 55 | 0 | 45 | +2 | 48 | 20 | 20.93 | 0.93 |
| 15. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| 16. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| 17. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| 18. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| 19. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| 20. | 0 | 55 | 0 | 45 | 0 | 24 | 26 | 26.22 | 0.22 |
| Effect | SS | MS | F | p-Value |
|---|---|---|---|---|
| “Var1” | 4.1329 | 4.1329 | 2.6980 | 0.13150 |
| “Var1^2” | 14.5746 | 14.5746 | 9.5146 | 0.01155 |
| “Var2” | 5.9065 | 5.9065 | 3.8558 | 0.07796 |
| “Var2^2” | 19.7532 | 19.7532 | 12.895 | 0.00492 |
| “Var3” | 26.4558 | 26.4558 | 17.270 | 0.00196 |
| “Var3^2” | 40.0032 | 40.0032 | 26.114 | 0.00045 |
| “Var1”*“Var2” | 8.0000 | 8.0000 | 5.2225 | 0.04537 |
| “Var1”*“Var3” | 0.5000 | 0.5000 | 0.3264 | 0.58039 |
| “Var2”*“Var3” | 4.5000 | 4.5000 | 2.9376 | 0.11730 |
| Source | SS | DF | MS | F- Value | Prob (p) |
|---|---|---|---|---|---|
| Whole model | 92.48 | 9 | 10.27 | 6.70 | 0.0031 |
| Residual | 15.31 | 10 | 1.53 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, N.; Singh, V.; Pandey, A.K.; Mishra, B.N.; Kulsoom, M.; Dasgupta, N.; Khan, S.; El-Enshasy, H.A.; Haque, S. Preparation and Evaluation of the ZnO NP–Ampicillin/Sulbactam Nanoantibiotic: Optimization of Formulation Variables Using RSM Coupled GA Method and Antibacterial Activities. Biomolecules 2019, 9, 764. https://doi.org/10.3390/biom9120764
Sharma N, Singh V, Pandey AK, Mishra BN, Kulsoom M, Dasgupta N, Khan S, El-Enshasy HA, Haque S. Preparation and Evaluation of the ZnO NP–Ampicillin/Sulbactam Nanoantibiotic: Optimization of Formulation Variables Using RSM Coupled GA Method and Antibacterial Activities. Biomolecules. 2019; 9(12):764. https://doi.org/10.3390/biom9120764
Chicago/Turabian StyleSharma, Nidhi, Vineeta Singh, Asheesh Kumar Pandey, Bhartendu Nath Mishra, Maria Kulsoom, Nandita Dasgupta, Saif Khan, Hesham A. El-Enshasy, and Shafiul Haque. 2019. "Preparation and Evaluation of the ZnO NP–Ampicillin/Sulbactam Nanoantibiotic: Optimization of Formulation Variables Using RSM Coupled GA Method and Antibacterial Activities" Biomolecules 9, no. 12: 764. https://doi.org/10.3390/biom9120764
APA StyleSharma, N., Singh, V., Pandey, A. K., Mishra, B. N., Kulsoom, M., Dasgupta, N., Khan, S., El-Enshasy, H. A., & Haque, S. (2019). Preparation and Evaluation of the ZnO NP–Ampicillin/Sulbactam Nanoantibiotic: Optimization of Formulation Variables Using RSM Coupled GA Method and Antibacterial Activities. Biomolecules, 9(12), 764. https://doi.org/10.3390/biom9120764