Abstract
Antimicrobial peptides (AMPs) are considered as a promising agent to overcome the drug-resistance of bacteria. Large numbers of AMPs have been identified from the skin secretion of Rana pipiens, including brevinins, ranatuerins, temporins and esculentins. In this study, the cDNA precursor of a broad-spectrum antimicrobial peptide, ranatuerin-2Pb, was cloned and identified. Additionally, two truncated analogues, RPa and RPb, were synthesised to investigate the structure-activity relationship of ranatuerin-2Pb. RPa lost antimicrobial activity against Candida albicans, MRSA, Enterococcus faecalis and Pseudomonas aeruginosa, while RPb retained its broad-spectrum antimicrobial activity. Additionally, ranatuerin-2Pb, RPa and RPb demonstrated inhibition and eradication effects against Staphylococcus aureus biofilm. RPb showed a rapid bacterial killing manner via membrane permeabilization without damaging the cell membrane of erythrocytes. Moreover, RPb decreased the mortality of S. aureus infected Galleria mellonella larvae. Collectively, our results suggested that RPb may pave a novel way for natural antimicrobial drug design.
1. Introduction
In recent decades, the overuse of conventional antibiotics has aggravated the resistance of bacteria, and this has become a serious problem facing the world [1]. Facing this severe problem, the World Health Organization (WHO) delivered a global report in 2014, which mentioned that a large number of bacteria possess drug resistance. Consequently, developing a global intervention plan against antimicrobial resistance is a matter of high urgency [2]. Therefore, it is urgent and critical to develop a new class of antibiotics. Nowadays, it is widely recognised that AMPs could pave a novel way to fight against multidrug-resistance strains because of the rapid kill-time of strains through cell membrane disruption, but the development of AMP-resistant strains could be inevitable when applied to clinical pathogens [3,4,5,6].
Ranatuerin-2 peptides, were initially identified in the skin of Lithobates catesbeianus and have been found in most species of North American, Chinese and Japanese frogs [7,8]. These peptides contain a C-terminal cyclic domain; however, the primary structure of these peptides was poorly conserved with several residue deletions with only the cysteine residues invariant. For instance, two peptides, ranatuerin-2P and ranatuerin-2Pa, which were isolated in Lithobates pipiens, consisted of a C-terminal cyclic heptapeptide domain and a “Rana box”, respectively [9]. The primary structures of pranatuerin-2 peptides were poorly conserved, which results in huge characteristic differences of antimicrobial properties and haemolytic activities. For instance, ranatuerin-2CHb possessed broad-spectrum antimicrobial activity [10], however, ranatuerin-2ONa exhibited the growth inhibition ability against Escherichia coli (E. coli) and Candida albicans (C. albicans) but was inactive against S. aureus [9].
In this study, the precursor encoding cDNA sequence of ranatuerin-2Pb was determined by the “shotgun” cloning method. In addition, two analogues, RPa (removed RT) and RPb (recited 18 C-terminal amino acids and amidated) were designed to investigate structure–activity relationship of ranatuerin-2Pb. Both ranatuerin-2Pb and its analogues were synthesised and the purified replicates were subjected to bioactivity assessments. Afterwards, the in vivo study was performed by waxworm model to investigate the anti-S. aureus effect of RPb.
2. Materials and Methods
2.1. Specimen Preparation and Secretion Harvesting
The specimens of the frog Rana pipiens were captured, settled and skin secretion was obtained from the dorsal skin as described previously [11]. The study was performed according to the guidelines in the UK Animal (Scientific Procedures) Act 1986, project license PPL 2694, issued by the Department of Health, Social Services and Public Safety, Northern Ireland. Procedures had been vetted by the IACUC of Queen’s University Belfast, and approved on 1st March, 2011.
2.2. “Shotgun” Cloning of a Novel Ranatuerin-2Pb Encoded cDNA Library
The “shotgun” cloning was carried out as previously described [11]. A NUP primer and a sense degenerated primer (5′-GAWYYAYYHRAGCCYAAADATG-3′; W = A + T, Y = C + T, H = A + C + T, R = A + G, D = A + G + T) were subjected to 3′-RACE reaction, from which the products were cloned and subsequently sequenced.
2.3. Peptide Synthesis
The novel peptide ranatuerin-2Pb (SFLTTVKKLVTNLAALAGTVIDTIKCKVTGGCRT) and two analogues RPa (SFLTTVKKLVTNLAALAGTVIDTIKCKVTGGC) and RPb (SFLTTVKKLVTNLAAL-NH2) were synthesized and purified by Gene Script Corporation (Nanjing, China) through solid-phase synthesis methods. The formation of disulfate bond was achieved by air-oxidation at room temperature for three days. The purity of the peptides was determined to be more than 95% by reverse-phase high-performance liquid chromatography (RP-HPLC). The peptides were further subjected to electrospray mass spectrometry to confirm their molecular mass.
2.4. Conformation Study
Physiochemical properties of helical wheel projection of all peptides were predicted by Heliquest (http://heliquest.ipmc.cnrs.fr). The secondary structures of peptides were evaluated by circular dichroism (CD) spectroscopy (JASCO J-815, Essex, UK), as described previously [12]. The peptides solutions were prepared at 50 µM in deionised water, 50% 2,2,2-trifluoroethanol (TFE)/H2O and large unilamellar liposome vesicles (LUVs) solutions.
DOPC, DOPE and DOPG (Avanti, Alabaster, AL, USA) were dissolved in chloroform to obtain stock solutions. The desired compositions were mixed and then dried under high vacuum evaporator until the lipid layer was formed at the bottom of the bottle. The resulting dispersions were extruded through a stack of two polycarbonate filters (200-nm pore size; Millipore Corp., Bedford, MA, USA) using a Liposofast low pressure homogeniser (Avestin, Ottawa, Canada) to obtain LUVs after five frozen-thaw cycles in 37°C. The concentration of total phosphorus was determined by ascorbic acid/ammonium molybdate method and 20 mM of liposomes was subjected to CD analysis. The estimated secondary structure was calculated by BESTSEL CD spectrum analysis tool [13].
2.5. Antimicrobial Activity
Antimicrobial activity assay was evaluated by confirmation of minimal inhibitory concentration (MIC) using two strains of Gram-negative bacterium, E. coli (NCTC 10418) and Pseudomonas aeruginosa (P. aeruginosa, ATCC 27853), three strains of Gram-positive bacterium, Staphylococcus aureus (NCTC 10788), Meticillin-resistant Staphylococcus aureus (MRSA, ATCC 12493) and Enterococcus faecalis (E. faecalis, NCTC 12697) and the yeast, C. albicans (NCPF 1467), as indicated previously [14]. 5 × 105 CFU/mL of bacterial suspension incubated with 2-fold dilution peptides (ranging from 1 µM to 256 µM) for 24 h at 37 °C. The antimicrobial activity of peptide was tested by monitoring the absorbance at 550nm using an ELISA plate reader (Biolise BioTek EL808, Winooski, VT, USA)
2.6. Antibiofilm Assays with Different Organisms
The antibiofilm ability of designed peptides was evaluated using two different organisms which were S. aureus (NCTC 10788), E. coli (NCTC 10418), as indicated previously [15]. For the MBIC assay, 5 × 105 CFU/mL bacterial cultures were incubated with 2-fold dilutions of peptides (ranging from 1 µM to 256 µM) for 24 h. Fresh medium and bacterial suspension as negative control and positive control, respectively. The absorbance values were measured at wavelength 595 nm using a Synergy HT plate reader (Biotech, Minneapolis, MN, USA). Experiments were done in triplicate and repeated twice independently.
2.7. Haemolysis Assay
The haemolysis assay was assessed using erythrocytes which were prepared from whole horse blood (TCS Biosciences Ltd. Buckingham, UK) as described previously [14]. PBS and 1% Triton-100 as negative and positive control, respectively. To further investigate the cell selectivity of the peptides, therapeutic index (TI) was calculated as HC50 divided by geometric mean of the peptide MICs against the relevant bacteria.
2.8. Membrane Permeability Assay
The membrane permeability assay was evaluated by SYTOXTM green nucleic acid stain (Life Technologies) as described previously [16]. The nucleic acid dye bound to cell nucleus when the cell membrane was permeabilised. The fluorescent intensity was detected with a Synergy HT plate reader (Biotech, Minneapolis, MN, USA) by an excitation and emission wavelength of 485 and 528 nm, respectively. S. aureus treated with melittin (8 µM) for 1 h were used as positive controls.
2.9. MTT and Lactate Dehydrogenase (LDH) Cytotoxicity Assay
The cell viability of ranatuerin-2Pb, RPa and RPb (Concentration ranging from 10-9-10-4) was analysed using 5 human cancer cell lines which are H157 (ATCC CRL-5802), MCF-7 (ATCC HTB-22), U251MG (ECACC General Cell Collection: 09063001), PC-3 (ATCC CRL-1435) and MDA-MB-435s (ATCC HTB-129). Melittin (Concentration ranging from 10−9–10−4) regarded as positive control. The cell viability was assessed using MTT assay as described previously [14]. The LDH cytotoxicity assay was performed using Pierce LDH Cytotoxicity Assay Kit (Thermo scientific, Rockford, IL, USA). The release of LDH was measured at 490 nm by a Synergy HT plate reader (Biotech, Minneapolis, MN, USA) after a 6 h incubation.
2.10. Time-Kill Assay
Bacterial suspensions were prepared as described above. The killing kinetics assay of designed peptides was evaluated as described before, with a little modification [17]. Bacteria were incubated with a dose of peptides corresponding to 1 × MIC and 4 × MIC. The viable count was monitored for up to 24 h. Aliquots of 10 µl were taken at different times and appropriately diluted in PBS buffer (pH 7.4), and spread onto MHA plates. The number of CFU were counted after incubating at 37 °C overnight. Three experiments were run in triplicate.
2.11. Assessing Efficacy of Peptides Against MRSA In Vivo
This in vivo study of RPb was assessed on waxworms (Galleria mellonella) as the previous study with minor modifications [18]. 250 ± 25 mg of the waxworms (Livefood UK Ltd., Rooks Bridge, UK) were inoculated with 10 µL MRSA bacteria suspension in autoclaved saline (108 CFU/mL). All the infected waxworms were confirmed to be alive at 2 h post-inoculation, followed by intrahaemocoelic injection of 20 mg/kg vancomycin (positive control group), 25 mg/kg peptides (sample group) and 50 mg/kg peptides (sample group). Saline and vancomycin (Sigma-Aldrich, Gillingham, UK) as negative control and positive control, respectively. Each group contained 9 waxworms and was repeated three times. All waxworms were observed every 12 h for 120 h.
2.12. Statistical Analysis
Data was analysed using Prism (Version 6.0; GraphPad Software Inc., San Diego, CA, USA). Error bars in the graphs represent standard error of the mean (SEM) with experiments performed on more than five replicates. Log-rank test was used to analyse the survival rate of waxworms. ∗ (p < 0.05), ∗∗ (p < 0.01) and ∗∗∗ (p < 0.001) represent significant difference.
3. Results
3.1. The Translated Open-Reading Frame Amino Acids Sequences of Ranatuerin-2 Peptides
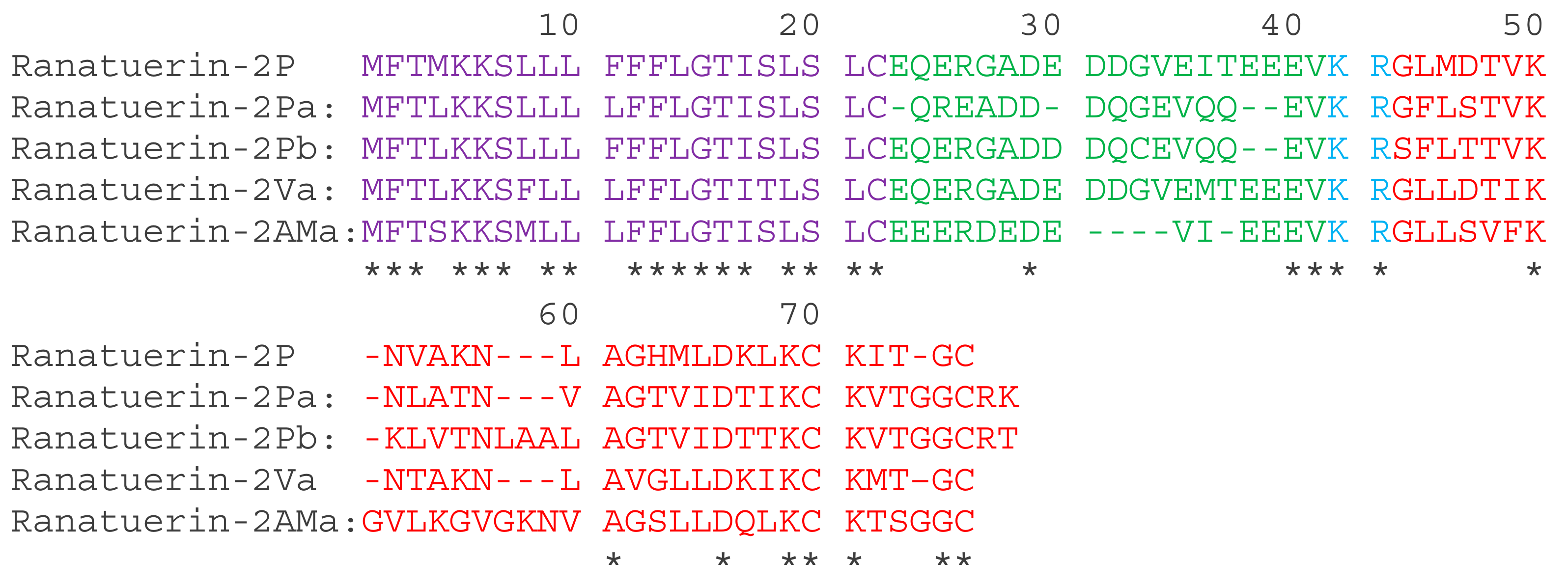
A full-length cDNA, encoding ranatuerin-2Pb, was successfully cloned from the skin secretion –derived cDNA library of Rana pipiens (Figure S1). The alignments of amino acid sequences of selected ranatuerin precursors are shown in Figure 1. There were several typical structural characteristics, including a putative signal peptide region of 22 amino acid residues, an acidic spacer peptide region, a classical -KR- propeptide convertase processing site and a mature peptide of 34 amino acid residues. The nucleotide sequence of ranatuerin-2Pb precursor was deposited in the Genbank Nucleotide Sequence Database under the accession number MK922296.
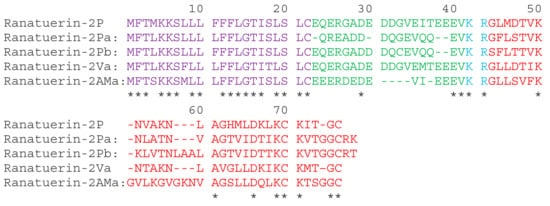
Figure 1.
The translated open-reading frame amino acids sequences of ranatuerin-2 peptides from different species of frogs. The sequences of mature peptides in are labelled in red. The sequences of signal peptide are labelled in purple. Asterisks indicate the identical amino acid residues. The acid spacer peptide regions are labelled in green. The processing sites of the precursor for releasing mature peptides are labelled in blue. Gaps (dashed line) were introduced to optimise the identities.
3.2. Peptide Design
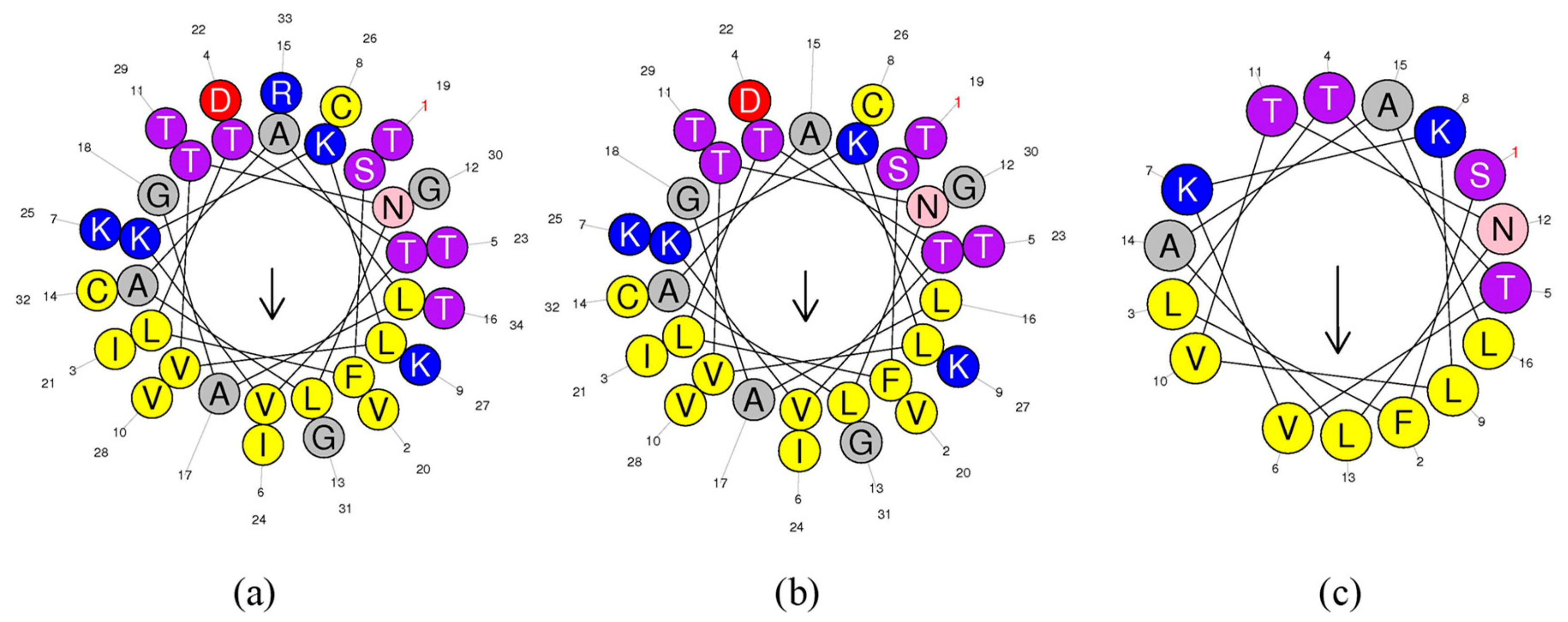
The helical wheel plots and secondary structures were predicted by online analysis tools. As shown in Figure 2, ranatuerin-2Pb and RPa shared a similar FVLGVIAVVLI hydrophobic face, however, RPb has less hydrophobic amino acid residues distributed on the hydrophobic face. Additionally, as shown in Table 1, ranatuerin-2Pb possessed +4 net charge with the hydrophobicity of 0.486 and the hydrophobic moment of 0.368; however, both RPa and RPb carried +3 net charge. Compared with ranatuerin-2Pb, RPa and RPb showed higher hydrophobicity which were 0.540 and 0.613, respectively (Table 1). All the peptides were purified by RP-HPLC and analysed by mass spectrometry (Figure S2).
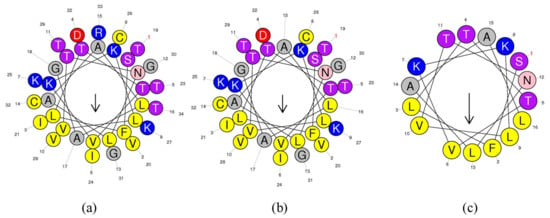
Figure 2.
Heliquest of peptides. Heliquest of (a) ranatuerin-2Pb, (b) RPa and (c) RPb.

Table 1.
Physio-chemical properties of Ranatuerin-2Pb, RPa and RPb calculated by HeliQuest online tool.
3.3. Secondary Structure Analysis
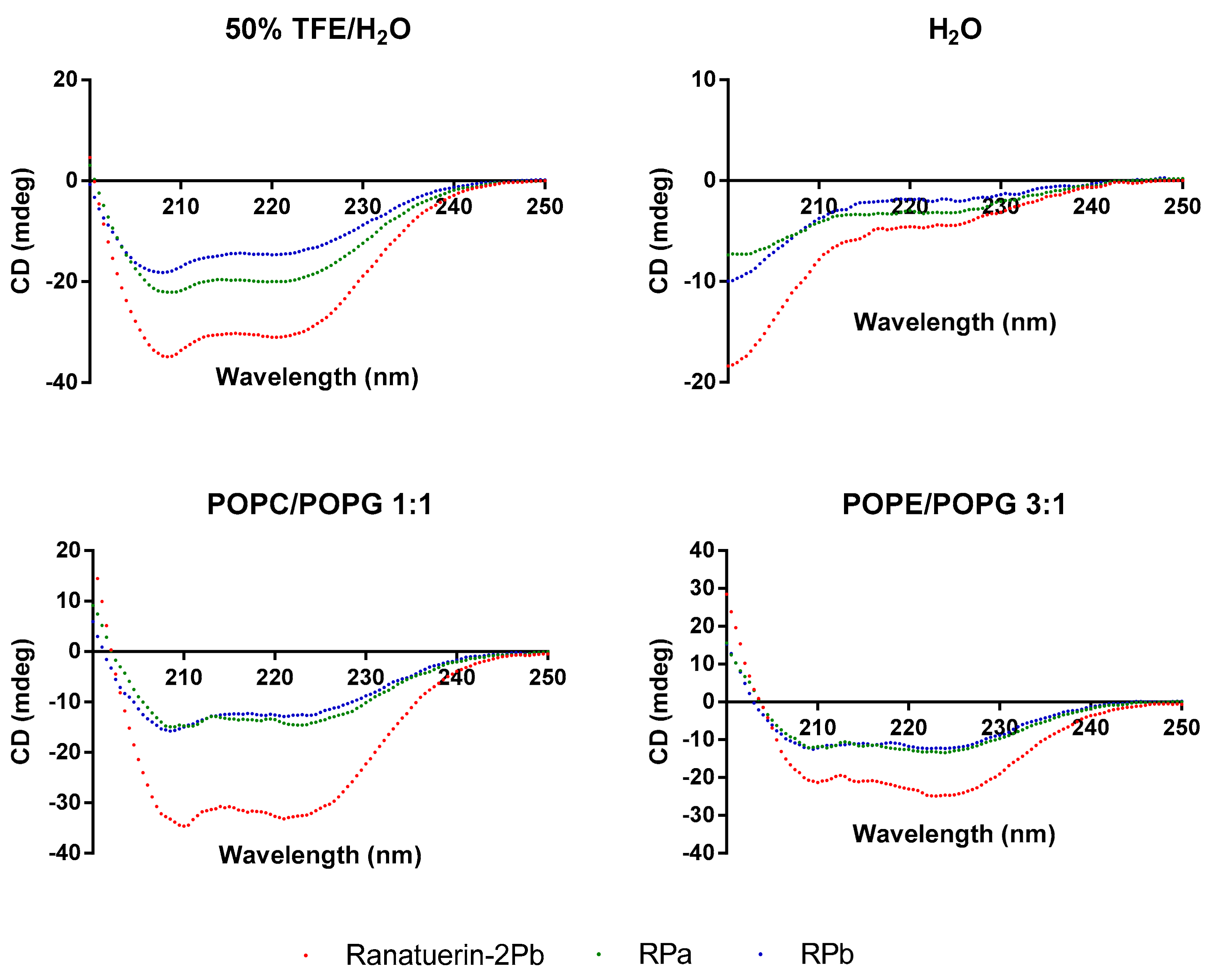
To demonstrate the changes of secondary structure of synthetic peptides in different environment, CD spectra was recorded in helical propagating solution (50% TFE/H2O), aqueous solution (H2O) and lipid bilayer liposomes of S. aureus (POPC/POPG 1:1) and E. coli (POPE/POPG 3:1) (Figure 3). All peptides were able to form α-helical structure in 50% TFE/H2O and membrane mimicking liposomes; however, the helical domain was eliminated in the aqueous environment. Based on the estimated secondary structure contents in Table 2, Ranatuerin-2Pb and RPb have similar helix proportion that are higher than RPa. Meanwhile, the helical content of Ranatuerin-2Pb was decreased when interacting with E. coli cell membrane-mimicking bilayer.
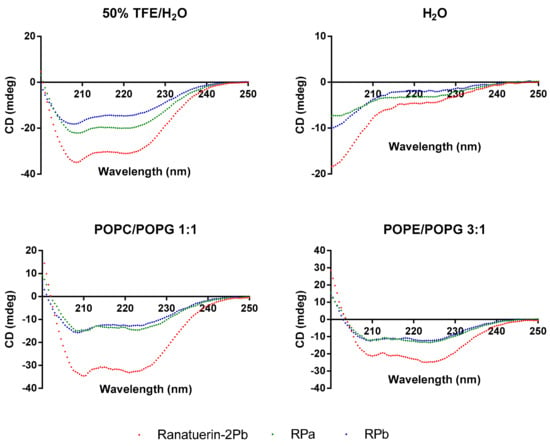
Figure 3.
CD spectra record for Ranatuerin-2Pb, RPa and RPb in 50% TFE/H2O, H2O and LUV liposomes of S. aureus (POPC/POPG 1:1) and E. coli (POPE/POPG 3:1).

Table 2.
The estimated secondary structure content (%) of Ranatuerin-2Pb, RPa and RPb from obtained CD spectra in different solutions using BESTSEL CD spectrum analysis tool.
3.4. Antimicrobial and Haemolytic Activity of Peptides
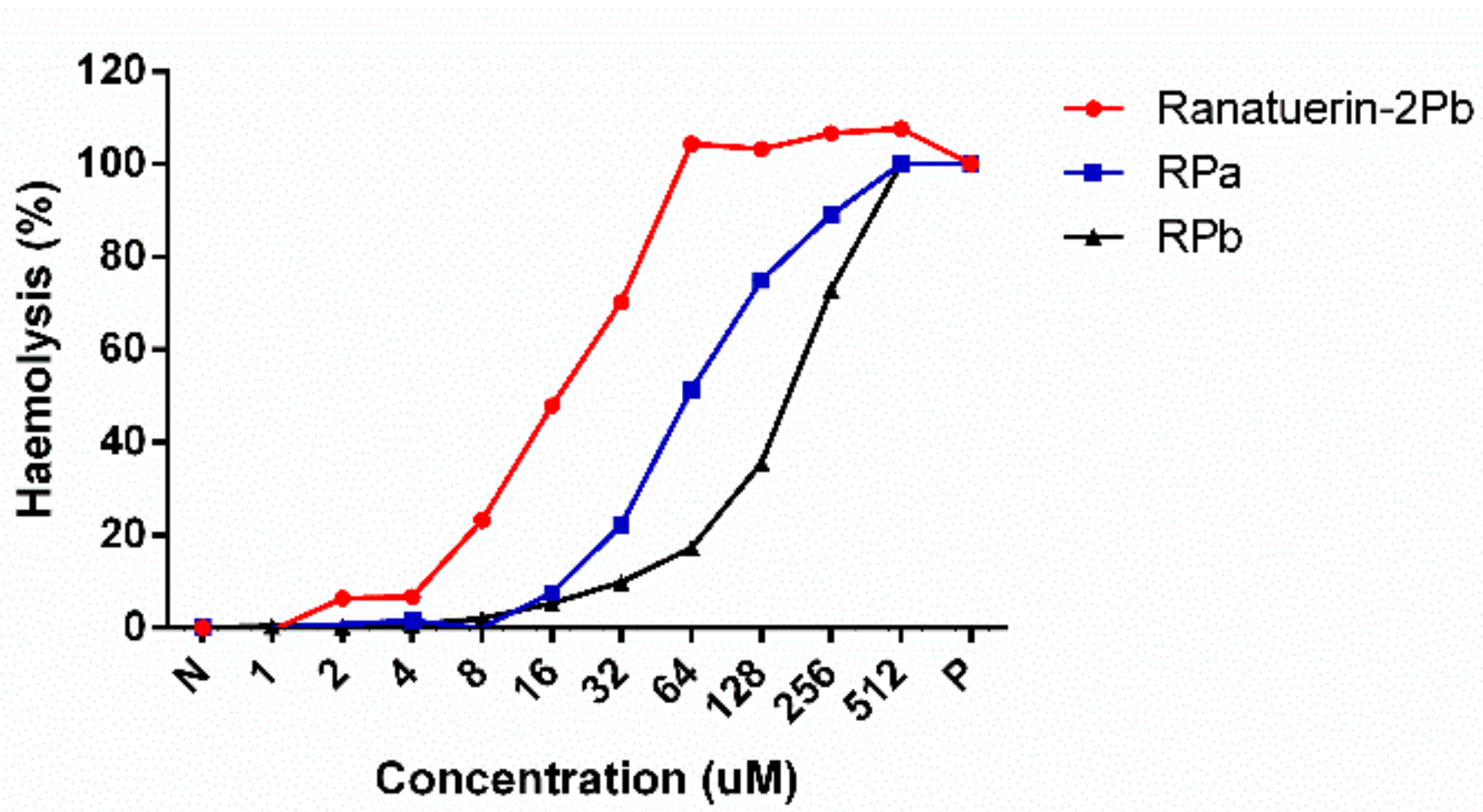
As shown in Table 3, ranatuerin-2Pb, RPa and RPb exhibited broad-spectrum antimicrobial activity. Specifically, ranatuerin-2Pb exhibited strong antimicrobial activity against S. aureus, E. coli, C. albicans and MRSA; however, it showed no inhibition activity against E. faecalis and P. aeruginosa. Additionally, RPa just possessed activity against S. aureus and E. coli. Notably, RPb exhibited significant broad-spectrum antimicrobial activity against all select tested microorganisms. Ranatuerin-2Pb, RPa and RPb exhibited a hemolysis rate near 20% on horse erythrocytes with the concentration at 8, 32 and 64 µM, respectively, as shown in Figure 4. In addition, The HC50 of ranatuerin-2Pb, RPa and RPb was 16.11, 63.90 and 178.0 µM, respectively. Notably, RPb exhibited the highest therapeutic index (TI) among the peptides.

Table 3.
Minimum inhibition concentrations (MICs) (µM) and minimum bactericidal concentrations (MBCs) (µM) of peptides against tested bacterias.
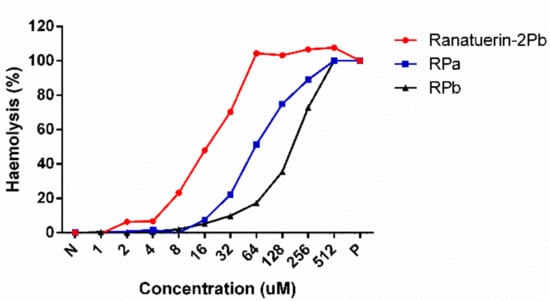
Figure 4.
Haemolytic activities of Ranatuerin-2Pb, RPa and RPb. PBS and 1% TritonX100 were employed as negative control (N) and positive control (P), respectively. The 100% haemolysis was achieved by 1% TritonX100.
3.5. Time-Kill Assay against S. aureus of Peptides
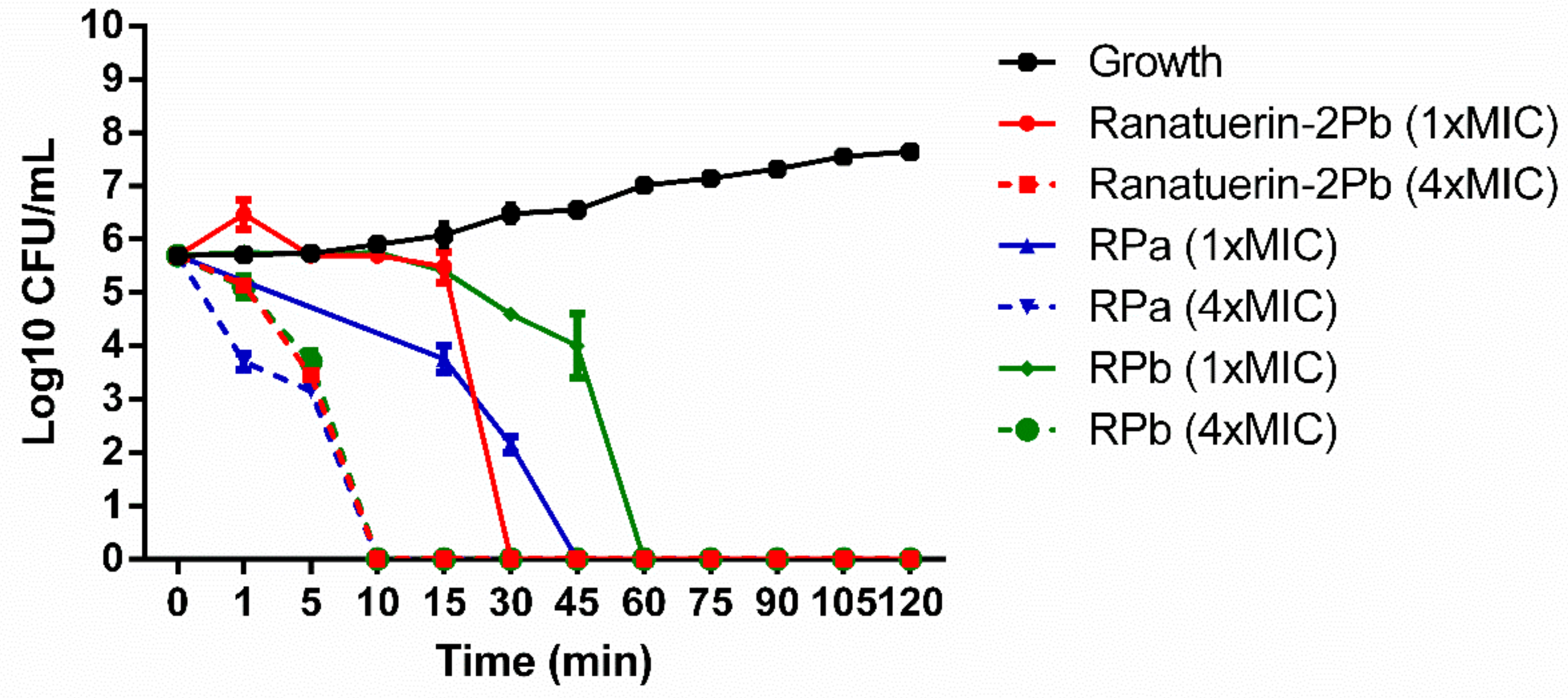
The killing effect of peptides against S. aureus was evaluated at 1 × MIC and 4 × MIC of peptides. As shown in Figure 5. The time-killing curve revealed that ranatuerin-2Pb, RPa and RPb at 1 × MIC exerted killing activity at 30, 45 and 60 min while they were able to kill all bacteria at 10 min at their 4 × MIC.
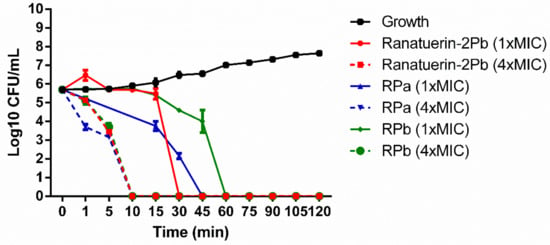
Figure 5.
Time-killing curve of S. aureus by ranatuerin-2Pb, RPa and RPb at different concentrations. Growth control correspond to bacteria incubated in PBS without peptide. Data represent means ± SEM of three independent experiments.
3.6. Antibiofilm Assay of Peptides against S. aureus
Ranatuerin-2Pb and RPb showed antibiofilm activity against S. aureus, E. coli and C. albicans. However, RPa just exhibited antibiofilm activity against S. aureus and E. coli biofilms. The minimum biofilm inhibitory concentration (MBIC) and the minimal biofilm eradication concentration (MBEC) of ranatuerin-2Pb, RPa and RPb against select tested bacteria biofilms are shown in Table 4.

Table 4.
Antibiofilm activity of peptides against tested bacteria.
3.7. MTT Cell Viability Assay
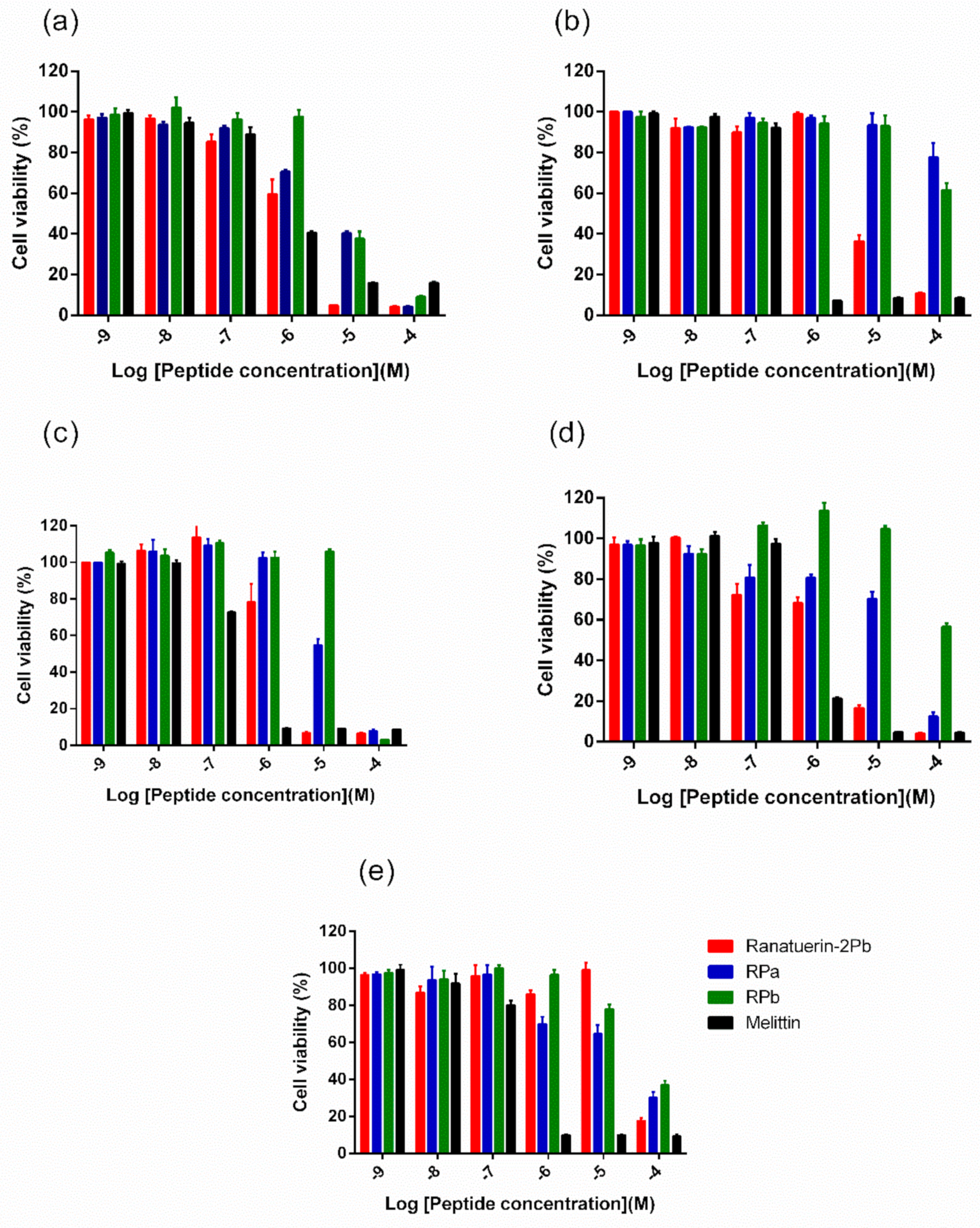
Five human cancer cell lines were used to evaluate the anticancer ability of peptides. As shown in Figure 6, ranatuerin-2Pb exhibited significant inhibitory effect on the proliferation of NCI-H157, MCF-7, U251MG and PC-3, with the IC50 value of 1.453 µM, 7.254 µM, 2.172 µM and 2.251 µM, respectively. However, RPa and RPb just possessed inhibitory effect on the proliferation of H157; the IC50 of RPa and RPb was 5.841 µM and 6.856 µM, respectively. In addition, ranatuerin-2Pb, RPa and RPb showed no inhibitory effect on the proliferation of MDA-MB-435s.
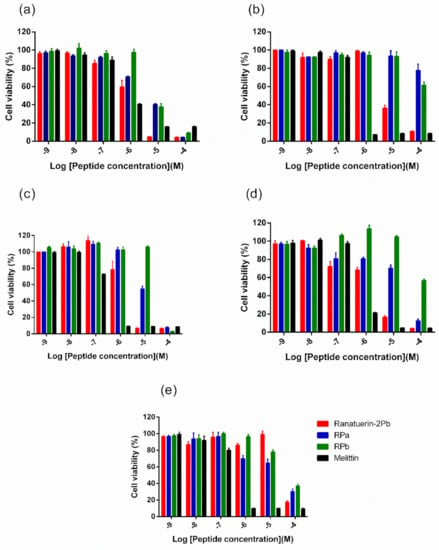
Figure 6.
The effect on cell proliferation of peptides on the cancer cell lines (a) NCI-H157, (b) MCF-7, (c) U251MG, (d) PC-3 and (e) MDA-MB-435s. Ranatuerin-2Pb, RPa, RPb and Melittin are labelled red, blue, green and black, respectively. The 100% cell viability was applied with growing cells without peptide treatment. Data points represent the average of three independent experiments with error bars presenting the SEM.
3.8. LDH Assay
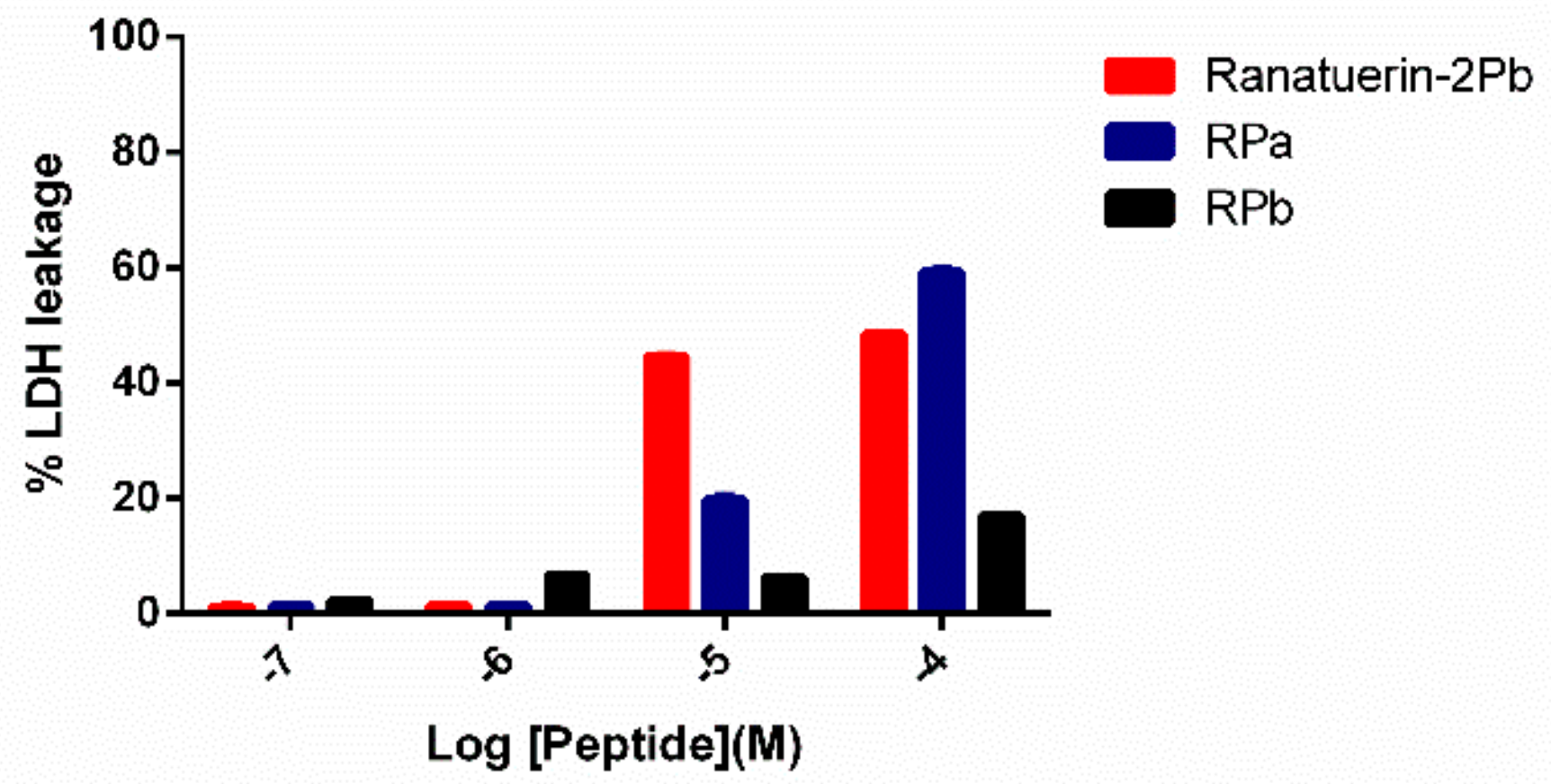
To further investigate the impact of ranatuerin-2Pb and its analogues on cancer viability, we examined their impact upon the lung cancer cell line NCI-H157 using the LDH assay. As shown in Figure 7, ranatuerin-2Pb induced the highest degree of LDH release for H157 among three peptides. However, RPb exhibited a negligible LDH leakage rate, which was consistent with the result of MTT assay.
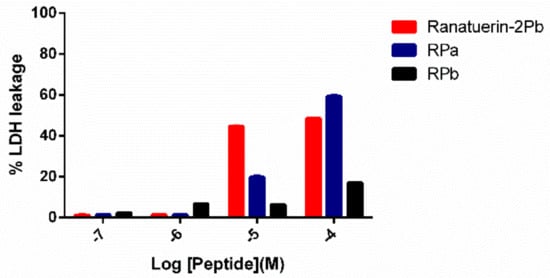
Figure 7.
LDH leakage in H157 cells after 30 min of treatment with peptides at different concentrations in serum-free medium. Ranatuerin-2Pb, RPa and RPb are labelled as red, blue and black, respectively.
3.9. Membrane Permeability Assay
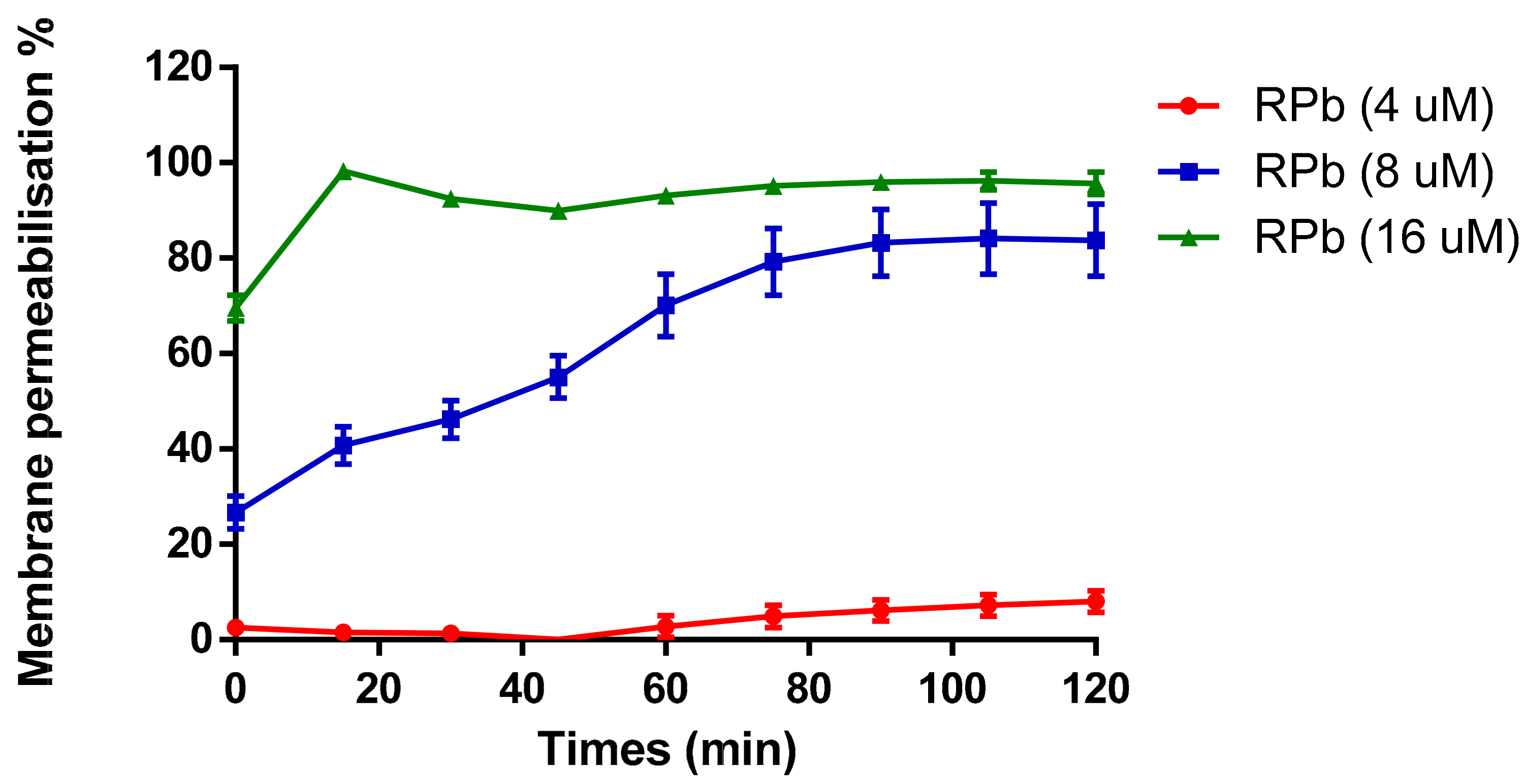
The cell membrane permeability of RPb was determined in a range of concentrations from 4 µM to 16 µM (Figure 8). It revealed that increasing the concentration of RPb induced the uptake of nucleus dye by S. aureus. With the increase of concentration of RPb, it induced significant and rapid membrane permeabilization. Moreover, 4×MIC of RPb resulted in an instant cell permeabilization effect that was similar to the effect of positive control, Melittin.
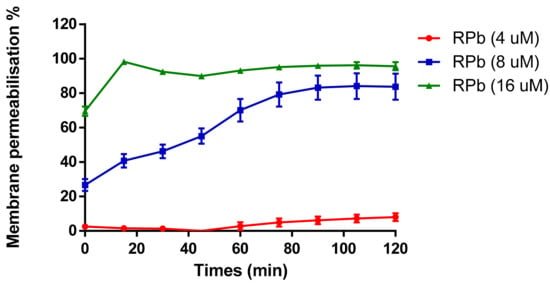
Figure 8.
Membrane permeability against S. aureus with addition of different concentrations of RPb. The 100% cell membrane permeabilisation effect was performed using 8 µM of Melittin. Data are the means ± standard errors of the means (n = 3).
3.10. Treatment of S. Aureus-Infected Waxworms with Peptides
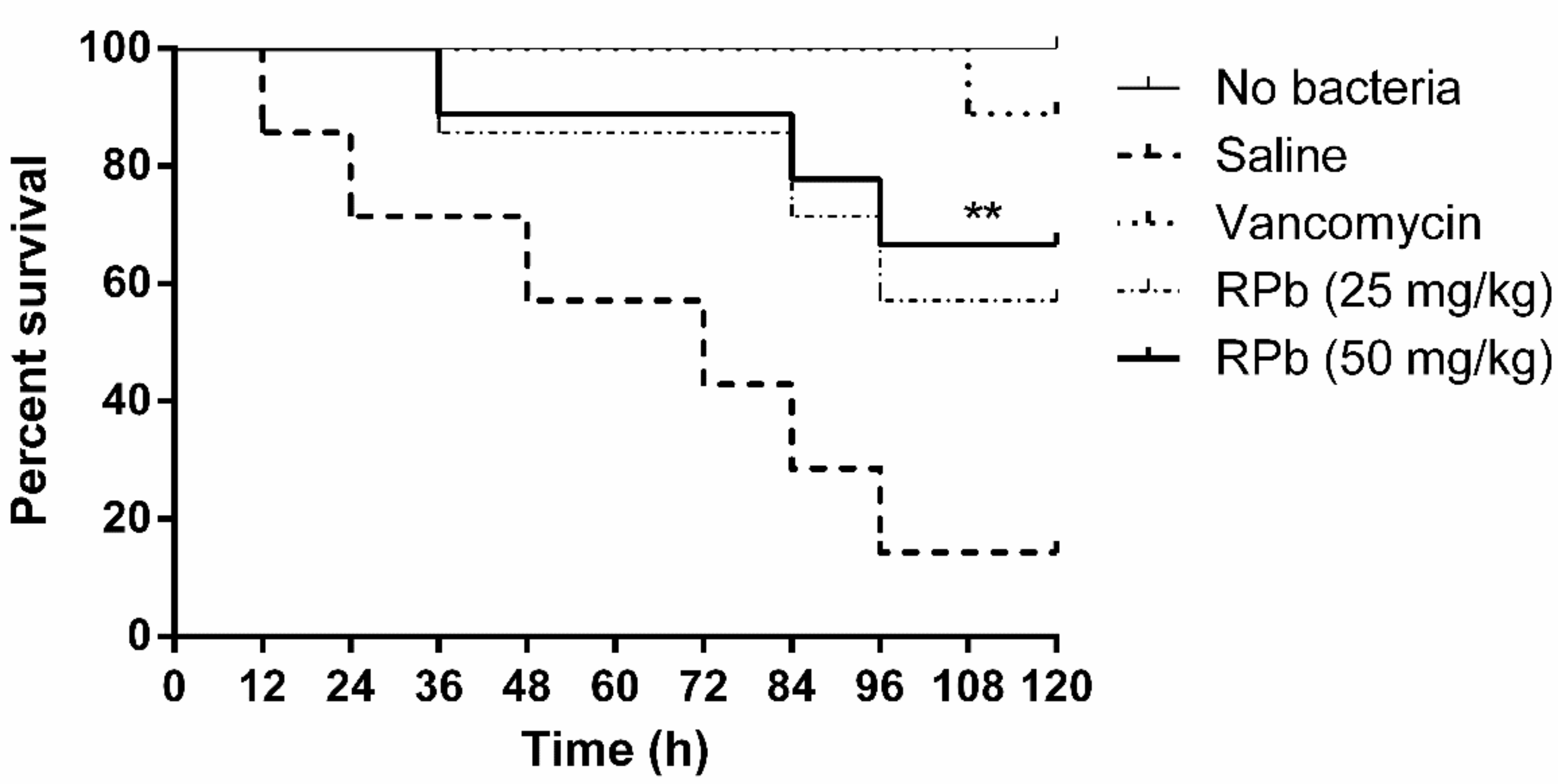
The mortality of S. aureus-infected waxworms after peptide treatment was significantly reduced as shown in Figure 9. Specifically, the survival rate of waxworms treated with 50 mg/kg of RPb was higher than that of waxworms treated with 25 mg/kg of RPb. In addition, RPb showed no cytotoxicity for waxworms.
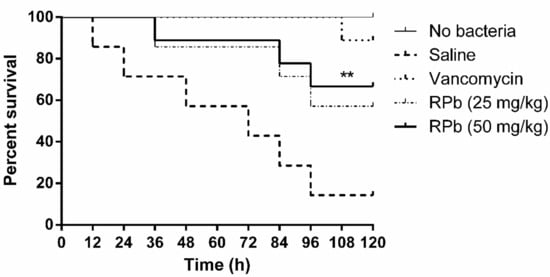
Figure 9.
The percentage survival waxworms infected with MRSA. The infected waxworms were treated with vancomycin (20 mg/kg) and RPb (25 mg/kg and 50 mg/kg), respectively. ** p < 0.01 and * p < 0.05 indicate significant difference.
4. Discussion
Ranatuerin-2 peptides have been found in most species of North American and Eurasian frogs [19], and it presented a less convincing phylogenetic marker than in the case of brevinin-1 and brevinin-2 [20]. Although ranatuerin-2Pb was previously identified in Rana pipiens [21], the cDNA encoding sequencing of this peptide was still unknown. Our study succeeded at the identification of the encoding gene of this skin defence peptide in the skin secretion of Rana pipiens. Recently, amounts of ranatuerin-2 peptides were isolated from skin secretions of Lithobates, odorrana and rana frogs [8]. Most ranatuerin-2 peptides possessed a broad-spectrum antimicrobial activity, though the primary sequences of these peptides are highly variable [20]. As shown in Figure 1, although there is a dramatic distinction in the length of mature peptide sequences and amino acid compositions, they still share common features, including the highly-conserved signal peptide sequence, the KR processing site and the C-terminal disulphate loop that exhibited unique sequence motif as -KCKXXGGC.
Recently, various principles were adopted in the design of peptides, such as the replacement of specific amino acids and the truncation of original peptides [22,23]. As we knew, net charge, hydrophobicity, amphipathicity and α-helicity of antimicrobial peptides play significant roles in terms of increasing antimicrobial activity and reducing cytotoxic effects [24]. Those characteristics facilitate the mechanisms that AMPs electrostatically attached on the cell membrane and further interact with the lipid layer. Nevertheless, a clear explanation is that basic residues in peptides contribute to binding the phospholipid head groups and then the membrane morphology could be changed by the formation of transient transmembrane pores that small molecules like ions and ATP could leak out of the cell [25].
Herein, we observed that ranatuerin-2Pb exhibited nonspecific killing effect against bacterial cells, cancer cells and even red blood cells. Such manner was also reported in the case of the AMPs, like melittin that can induce the detergent-like effect upon the cell membrane [26]. To our understanding, eliminating the cytotoxicity without sacrificing biofunction could be achieved through optimisation of the ratio of these characteristics [27]. As our study showed, the removal of last two residues of ranatuerin-2Pb contributed to the decrease of the helical content for RPa that could explain the reduction of antimicrobial, anticancer and haemolytic activity for RPa compared to the parent peptide (Table 2). Although there is no NMR analysis performed for ranatuerin-2 peptides, the studies on Gaegurin, a 32-mer AMP from Rana rugosa possessing a C-terminal loop, illustrates that the structure is depicted by two locally well-defined helical domain helices, which are composed of residues 2–10 and 16–32 [28]. It suggests that ranatuerin-2Pb might also possess two helical segments at respective N-terminus and C-terminus, and the last two residues are probably included in the helix domain. Therefore, the removal of C-terminal residues results in the decrease of helical content that weakens the membrane permeabilisation.
C-terminal “Rana box” is a unique structure among the AMPs discovered from Ranidae frogs. The function of the intramolecular disulphate bond has been considered to stably constrain the fold of C-terminal helical loop [29]. Functions of “Rana box” are remained ambiguous and controversial because the bioactivity of peptides changed or remained by removal of the loop were reported [30]. As our previous study demonstrated, the removal of C-terminal loop of nigrocin-HL increased the antimicrobial activity and, replaced by an amidated phenylalanine residue, resulted in the further enhancement of antimicrobial activity and reducing haemolysis activity [31]. Thus, further shortening the length of amino acids was performed. Notably, the helical content of RPb is around the similar proportion to RPa through the CD analysis (Table 2), which is consistent with our assumption about two helix segments located at both terminus of ranatuerin-2Pb. The N-terminal 16 amino acids could be the main contributors in the formation of α-helix at N-terminus.
Interestingly, the 16-mer N-terminal segment, RPb, not only restored and broadened the antimicrobial activity but also reduced the toxicity to mammalian cells (Table 3 and Figure 6). Even more, the truncation slowed down the killing effect against S. aureus. It is reported that the two helical domains of Gaegurin induced a hinge region near Gly24, which the N-terminal helix could bend and insert into bilayer to form transmembrane pore while the C-terminal helix could attach on the lipid surface [29]. Such a feature may lead to the detergent-like effect at very low peptide-to-lipid ratios for membrane disintegration [32]. Also, the cytolytic peptide, Melittin, formed a hinge near Pro14 that results in two helical segments; however, the removal of the hinge domain significantly decreased the haemolysis [33]. The similar results were observed herein for RPb, indicating that the helix–kink–helix structure could improve the general membrane permeabilisation effect without specificity. RPb still retains the helical domain for insertion into the cell membrane, but it might need higher peptide-to-lipid ratios for inducing apparent membrane disintegration, which could be proven by the instant membrane permeabilisation caused by 16 µM of RPb (Figure 8). With regards to the reason of broad-spectrum activity of RPb, it is though unclarified, we assume that it would be influenced by the affinity towards different lipids [34]. Due to the helix–kink–helix conformation in ranatuerin-2Pb and RPa, they may be easy to diffuse across the peptidoglycan matrix of Gram-positive bacteria first and then act on the cytoplasmic membrane; however, they may be challenging to diffuse across the outer membranes of Gram-negative bacteria, which results in the loss of antimicrobial activity [35]. Taken together, our results revealed that a delicate balance of amphipathicity and α-helical is necessary for broad-spectrum antimicrobial activity and less toxicity.
Bacteria biofilms, the self-produced polysaccharide matrix that facilitates the adherence of microorganisms, provide better protection for bacteria, which can lead to antibiotic resistance in clinical infections [36]. In particular, sessile bacteria could tolerate much higher concentration of antimicrobial agents than the planktonic bacteria [37]. Our results demonstrated that ranatuerin-2Pb, RPa and RPb exhibited potent inhibition on the growth of biofilm against S. aureus (Table 3). Although the mechanism is unclear, a theory that the AMPs are transferred through the pores formed by the binding of the extracellular biofilm or only completely dispersed in the biofilm may explain this phenomenon [38].
Apart from antimicrobial activity, some AMPs also exhibited outstanding anticancer effects. Our results revealed ranatuerin-2Pb, RPa and RPb exhibited different anticancer effects on various cancer cells (Figure 6). Several theories have tried to explain the anticancer mechanism, including the interaction between AMPs and tumour cells, cell apoptosis, caspase activation, cytochrome C release, DNA fragmentation and mitochondrial membrane depolarisation [39,40,41,42]. However, based on our hypothesis above and the results of the LDH assay, the anticancer mechanism may mainly involve disruption of the cell membrane [41].
RPb exhibited higher therapeutic index, we therefore investigated the antimicrobial activity of RPb in the S. aureus-infected Galleria mellonella larvae model [43]. Although RPb shows a significant effect against S. aureus in vivo, the efficacy is still lower than that of the lead antibiotics. A similar study also reported that the antimicrobial potency was less than that of vancomycin in the treatment of MRSA infection [44]. It might be the nature of poor stability of AMPs in vivo. Further studies on peptide stability or even bioavailability should be taken into consideration. Additionally, RPb showed no acute toxicity in waxworms, which could provide a reference for further in vivo study.
5. Conclusions
Ranatuerin-2Pb is an antimicrobial peptide that can form a helical structure to permeabilise bacterial cell membrane. It is potent against various microorganisms including the antibiotic-resistance strain, MRSA. However, the coherent helix–kink–helix structure may not be favourable to the selectivity towards bacteria and mammalian cell membrane. The truncated analogue, RPb, is able to enlarge the antimicrobial spectrum with the decrease of haemolytic effect. Although it exhibits significant antimicrobial activity both in vitro and in vivo, which warrants further investigation, detailed explanation of the mechanisms needs to be clarified. Overall, Ranatuerin-2Pb is a good antimicrobial template for further studying and developing new antimicrobial compounds.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/9/6/249/s1, Figure S1. Nucleotide and corresponding translated open-reading frame of precursor cDNA cloned from the skin secretion of Chinese piebald odorous frog cDNA library that encodes ranatuerin-2Pb. Figure S2. RP-HPLC chromatogram and mass spectrums of synthesised peptides, ranatuerin-2Pb (a), RPa (b) and RPb (c).
Author Contributions
Conceptualization, M.Z., L.W., T.C. and H.F.K. Formal analysis, C.M., X.X. and H.F.K. Funding acquisition, R.Z. and H.F.K. Investigation, X.Z. and D.S. Methodology, C.M. and X.X. Resources, D.S. and R.Z. Supervision, X.X. Validation, X.Z., Z.Y., M.Z. and X.X. Writing—original draft, X.Z. and D.S. Writing—review & editing, X.X., T.C. and H.F.K.
Funding
This work was funded by Science and Technology Planning Project of Guangdong Province [2015A010107018], Shaoguan Science and Technology Planning Project [2018sn157] and Shaoguan Social Development Science and Technology Plan Project (2018Sn080). H.F.K. was supported by the Science and Technology Development Fund of Macau SAR (FDCT) [019/2017/A1] and the Faculty of Health Sciences (FHS) University of Macau (UM). X. X. was in receipt of a Postdoctoral Fellowship from the FHS UM from 2015 to 2017.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A review on antibiotic resistance: Alarm bells are ringing. Cureus 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Organization, W.H. Antimicrobial Resistance Global Report on Surveillance: 2014 Summary; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Yan, J.; Wang, K.; Dang, W.; Chen, R.; Xie, J.; Zhang, B.; Song, J.; Wang, R. Two hits are better than one: Membrane-active and DNA binding-related double-action mechanism of nk-18, a novel antimicrobial peptide derived from mammalian nk-lysin. Antimicrob. Agents Chemother. 2013, 57, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef] [PubMed]
- Maria-Neto, S.; de Almeida, K.C.; Macedo, M.L.R.; Franco, O.L. Understanding bacterial resistance to antimicrobial peptides: From the surface to deep inside. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2015, 1848, 3078–3088. [Google Scholar] [CrossRef] [PubMed]
- Fleitas, O.; Agbale, C.M.; Franco, O.L. Bacterial resistance to antimicrobial peptides: An evolving phenomenon. Front Biosci. 2016, 21, 1013–1038. [Google Scholar]
- Goraya, J.; Knoop, F.C.; Conlon, J.M. Ranatuerins: Antimicrobial peptides isolated from the skin of the american bullfrog, rana catesbeiana. Biochem. Biophys. Res. Commun. 1998, 250, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, R. The chemistry and biological activities of peptides from amphibian skin secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef]
- Conlon, J.M.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; King, J.D. Primary structures of skin antimicrobial peptides indicate a close, but not conspecific, phylogenetic relationship between the leopard frogs lithobates onca and lithobates yavapaiensis (ranidae). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2010, 151, 313–317. [Google Scholar] [CrossRef]
- Conlon, J.M.; Mechkarska, M.; Coquet, L.; Jouenne, T.; Leprince, J.; Vaudry, H.; Kolodziejek, J.; Nowotny, N.; King, J.D. Characterization of antimicrobial peptides in skin secretions from discrete populations of lithobates chiricahuensis (ranidae) from central and southern arizona. Peptides 2011, 32, 664–669. [Google Scholar] [CrossRef]
- Zhou, X.; Ma, C.; Zhou, M.; Zhang, Y.; Xi, X.; Zhong, R.; Chen, T.; Shaw, C.; Wang, L. Pharmacological effects of two novel bombesin-like peptides from the skin secretions of chinese piebald odorous frog (odorrana schmackeri) and european edible frog (pelophylax kl. Esculentus) on smooth muscle. Molecules 2017, 22, 1798. [Google Scholar] [CrossRef]
- Wu, D.; Gao, Y.; Tan, Y.; Liu, Y.; Wang, L.; Zhou, M.; Xi, X.; Ma, C.; Bininda-Emonds, O.R.; Chen, T. Discovery of distinctin-like-peptide-ph (dlp-ph) from the skin secretion of phyllomedusa hypochondrialis, a prototype of a novel family of antimicrobial peptide. Front. Microbiol. 2018, 9, 541. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Dermaseptin-ph: A novel peptide with antimicrobial and anticancer activities from the skin secretion of the south american orange-legged leaf frog, pithecopus (phyllomedusa) hypochondrialis. Molecules 2017, 22, 1805. [Google Scholar] [CrossRef]
- Nair, S.; Desai, S.; Poonacha, N.; Vipra, A.; Sharma, U. Antibiofilm activity and synergistic inhibition of staphylococcus aureus biofilms by bactericidal protein p128 in combination with antibiotics. Antimicrob Agents Chemother 2016, 60, 7280–7289. [Google Scholar]
- Chen, Q.; Cheng, P.; Ma, C.; Xi, X.; Wang, L.; Zhou, M.; Bian, H.; Chen, T. Evaluating the bioactivity of a novel broad-spectrum antimicrobial peptide brevinin-1gha from the frog skin secretion of hylarana guentheri and its analogues. Toxins 2018, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Pag, U.; Oedenkoven, M.; Papo, N.; Oren, Z.; Shai, Y.; Sahl, H.-G. In vitro activity and mode of action of diastereomeric antimicrobial peptides against bacterial clinical isolates. J. Antimicrob. Chemother. 2004, 53, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Desbois, A.P.; Coote, P.J. Wax moth larva (galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef]
- Chen, T.; Zhou, M.; Rao, P.; Walker, B.; Shaw, C. The chinese bamboo leaf odorous frog (rana (odorrana) versabilis) and north american rana frogs share the same families of skin antimicrobial peptides. Peptides 2006, 27, 1738–1744. [Google Scholar] [CrossRef]
- Conlon, J.M.; Kolodziejek, J.; Nowotny, N. Antimicrobial peptides from ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim. Et Biophys. Acta (Bba)-Proteins Proteom. 2004, 1696, 1–14. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Blouin, M.S. Selection for antimicrobial peptide diversity in frogs leads to gene duplication and low allelic variation. J. Mol. Evol. 2007, 65, 605–615. [Google Scholar] [CrossRef]
- Zhu, X.; Dong, N.; Wang, Z.; Ma, Z.; Zhang, L.; Ma, Q.; Shan, A. Design of imperfectly amphipathic α-helical antimicrobial peptides with enhanced cell selectivity. Acta Biomater. 2014, 10, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Ong, Z.Y.; Wiradharma, N.; Yang, Y.Y. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv. Drug Deliv. Rev. 2014, 78, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.-C.; Strandberg, E.; Grau-Campistany, A.; Wadhwani, P.; Reichert, J.; Bürck, J.; Rabanal, F.; Auger, M.l.; Paquin, J.-F.o.; Ulrich, A.S. Influence of the length and charge on the activity of α-helical amphipathic antimicrobial peptides. Biochemistry 2017, 56, 1680–1695. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238. [Google Scholar] [CrossRef] [PubMed]
- Parachin, N.S.; Franco, O.L. New edge of antibiotic development: Antimicrobial peptides and corresponding resistance. Front. Microbiol. 2014, 5, 147. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, Y.K.; Park, J.W.; Lee, B.; Lee, B.J. Solution structure of the antimicrobial peptide gaegurin 4 by 1h and 15n nuclear magnetic resonance spectroscopy. Eur. J. Biochem. 2000, 267, 2695–2704. [Google Scholar] [CrossRef]
- Won, H.-S.; Kang, S.-J.; Lee, B.-J. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2009, 1788, 1620–1629. [Google Scholar] [CrossRef]
- Abraham, P.; Sundaram, A.; Asha, R.; Reshmy, V.; George, S.; Kumar, K.S. Structure-activity relationship and mode of action of a frog secreted antibacterial peptide b1ctcu5 using synthetically and modularly modified or deleted (smmd) peptides. PLoS ONE 2015, 10, e0124210. [Google Scholar] [CrossRef]
- Bao, K.; Yuan, W.; Ma, C.; Wang, L.; Hong, M.; Xi, X.; Zhou, M.; Chen, T. Modification targeting the “rana box” motif of a novel nigrocin peptide from hylarana latouchii enhances and broadens its potency against multiple bacteria. Front. Microbiol. 2018, 9, 2846. [Google Scholar] [CrossRef]
- Bechinger, B.; Lohner, K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, R.; Bhunia, A.; Bhattacharjya, S. Micelle-bound structures and dynamics of the hinge deleted analog of melittin and its diastereomer: Implications in cell selective lysis by d-amino acid containing antimicrobial peptides. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2010, 1798, 128–139. [Google Scholar] [CrossRef]
- Maturana, P.; Martinez, M.; Noguera, M.E.; Santos, N.; Disalvo, E.A.; Semorile, L.; Maffia, P.C.; Hollmann, A. Lipid selectivity in novel antimicrobial peptides: Implication on antimicrobial and hemolytic activity. Colloids Surf. B Biointerfaces 2017, 153, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane active antimicrobial peptides: Translating mechanistic insights to design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A. Controlling Biofilm and Persister Cells by Targeting Cell Membranes. Ph.D. Thesis, Syracuse University, Syracuse, NY, USA, 2015. [Google Scholar]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635. [Google Scholar] [CrossRef]
- Sok, M.; Šentjurc, M.; Schara, M.; Stare, J.; Rott, T. Cell membrane fluidity and prognosis of lung cancer. Ann. Thorac. Surg. 2002, 73, 1567–1571. [Google Scholar] [CrossRef]
- Lee, J.; Lee, D.G. Antimicrobial peptides (amps) with dual mechanisms: Membrane disruption and apoptosis. J. Microbiol. Biotechnol. 2015, 25, 759–764. [Google Scholar] [CrossRef]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zai, Y.; Xi, X.; Ma, C.; Wang, L.; Zhou, M.; Shaw, C.; Chen, T. A novel membrane-disruptive antimicrobial peptide from frog skin secretion against cystic fibrosis isolates and evaluation of anti-mrsa effect using galleria mellonella model. Biochim. Et Biophys. Acta (Bba)-Gen. Subj. 2019, 1863, 849–856. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).