Role of Transmembrane Proteins for Phase Separation and Domain Registration in Asymmetric Lipid Bilayers
Abstract
:1. Introduction
2. Theory
2.1. Free Energy of a Lipid Membrane That Contains Transmembrane Proteins
2.2. Spinodals and Critical Points
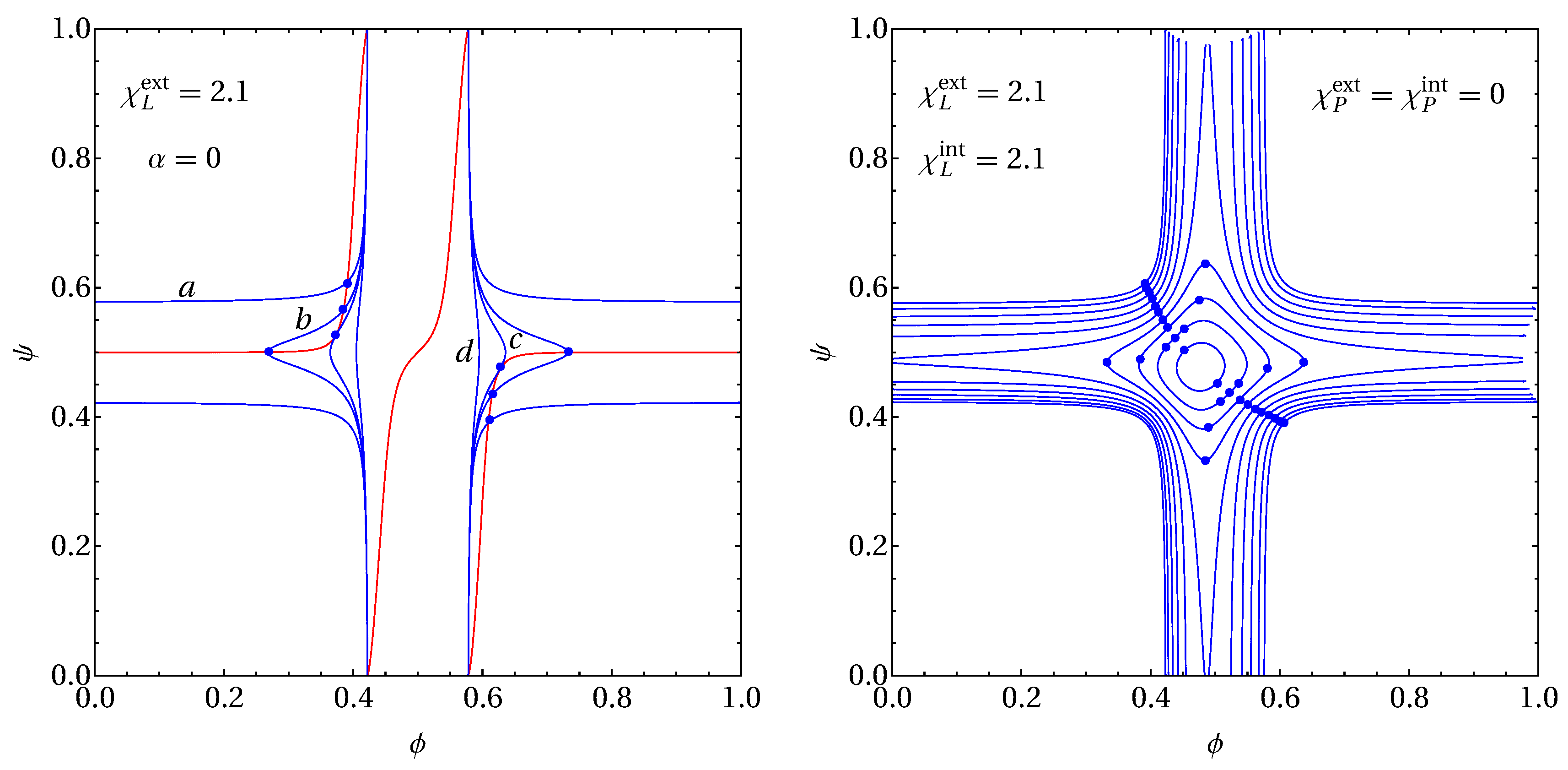
2.3. Example for Calculation of Spinodals and Critical Points
2.4. Numerical Calculation of Coexisting Phases
2.5. Free Energy of a Lipid Membrane That Contains Peripheral Proteins
2.6. A Simple Example for Phase Stability in the Presence of Transmembrane versus Peripheral Proteins
3. Results
3.1. Phase Behavior in the Absence of Proteins
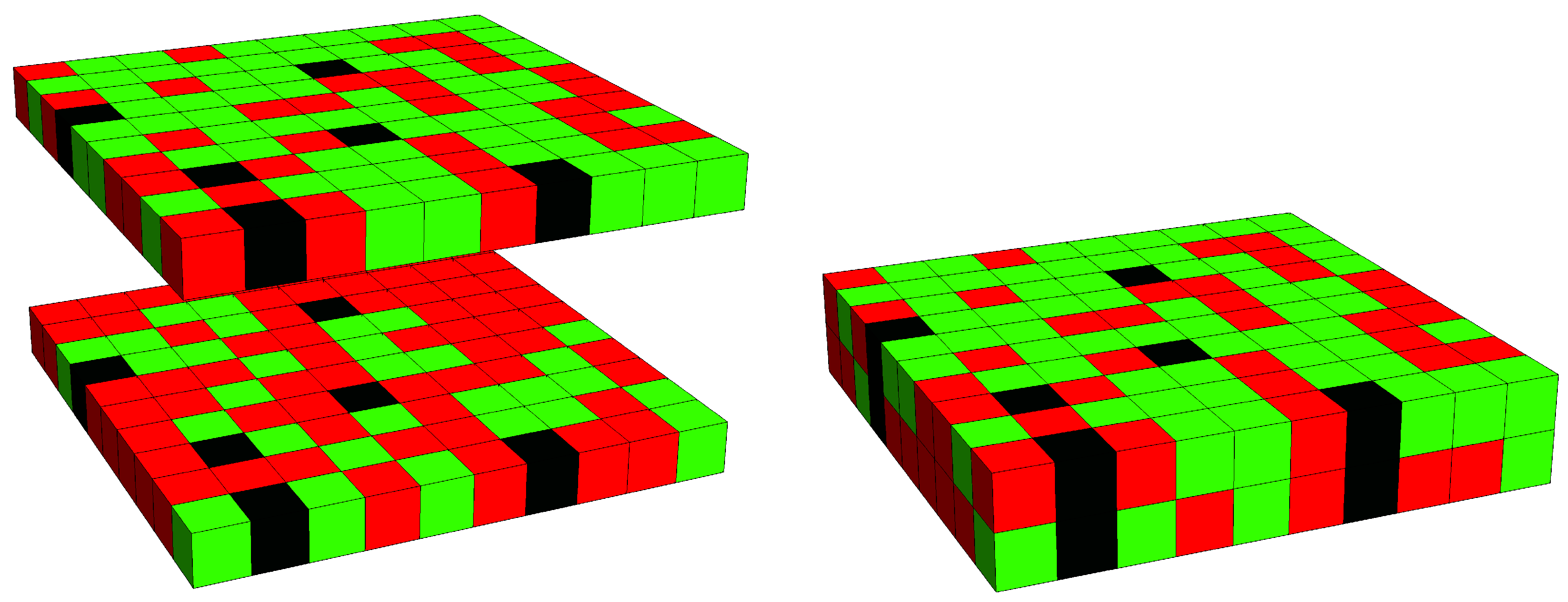
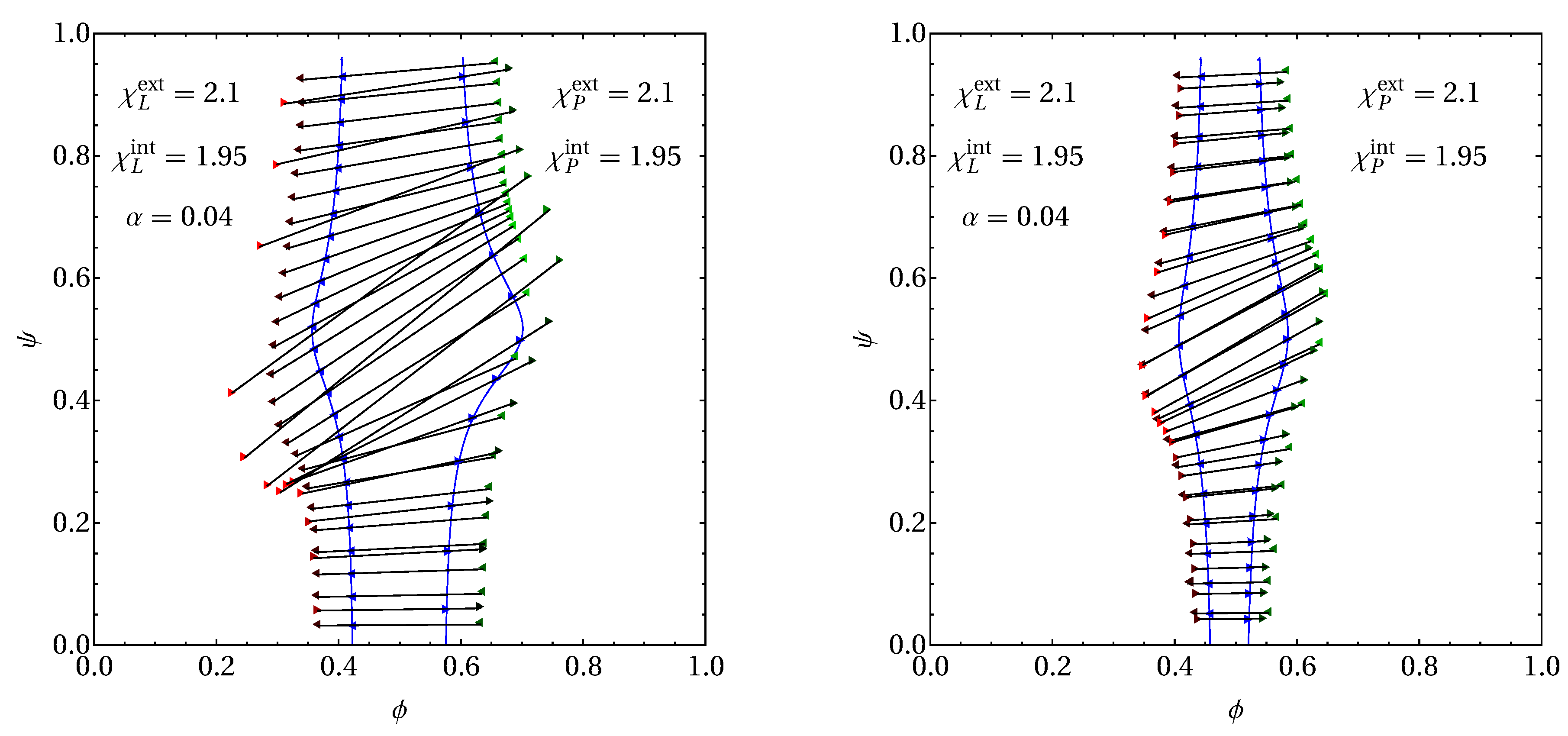
3.2. Phase Behavior in the Presence of Proteins
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A. Thermodynamic Criterion for Critical Point
References
- Almeida, P.F. Thermodynamics of lipid interactions in complex bilayers. Biochim. Biophys. Acta Biomembr. 2009, 1788, 72–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feigenson, G.W. Phase diagrams and lipid domains in multicomponent lipid bilayer mixtures. Biochim. Biophys. Acta Biomembr. 2009, 1788, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, P.; Baird, B.; Holowka, D. Lipid rafts, fluid/fluid phase separation, and their relevance to plasma membrane structure and function. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 18, pp. 583–590. [Google Scholar]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Sevcsik, E.; Schütz, G.J. With or without rafts? Alternative views on cell membranes. Bioessays 2016, 38, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Parmryd, I. Interleaflet coupling, pinning, and leaflet asymmetry—Major players in plasma membrane nanodomain formation. Front. Cell Dev. Biol. 2017, 4, 155. [Google Scholar] [CrossRef]
- Veatch, S.L.; Keller, S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003, 85, 3074–3083. [Google Scholar] [CrossRef]
- Veatch, S.L.; Keller, S.L. Seeing spots: Complex phase behavior in simple membranes. Biochim. Biophys. Acta Mol. Cell Res. 2005, 1746, 172–185. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.Y.; Silvius, J.R. Cholesterol does not induce segregation of liquid-ordered domains in bilayers modeling the inner leaflet of the plasma membrane. Biophys. J. 2001, 81, 2762–2773. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Sachs, J.N. Interleaflet interaction and asymmetry in phase separated lipid bilayers: Molecular dynamics simulations. J. Am. Chem. Soc. 2011, 133, 6563–6577. [Google Scholar] [CrossRef]
- Raghupathy, R.; Anilkumar, A.A.; Polley, A.; Singh, P.P.; Yadav, M.; Johnson, C.; Suryawanshi, S.; Saikam, V.; Sawant, S.D.; Panda, A.; et al. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 2015, 161, 581–594. [Google Scholar] [CrossRef]
- Levental, I.; Veatch, S.L. The continuing mystery of lipid rafts. J. Mol. Biol. 2016, 428, 4749–4764. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Vaz, W.L.C. Model systems, lipid rafts, and cell membranes. Ann. Rev. Biophys. Biomol. Struct. 2004, 33, 269–295. [Google Scholar] [CrossRef] [PubMed]
- Levental, K.R.; Levental, I. Giant plasma membrane vesicles: Models for understanding membrane organization. In Current Topics in Membranes; Elsevier: Amsterdam, The Netherlands, 2015; Volume 75, pp. 25–57. [Google Scholar]
- Bennett, W.D.; Tieleman, D.P. Computer simulations of lipid membrane domains. Biochim. Biophys. Acta Biomembr. 2013, 1828, 1765–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komura, S.; Andelman, D. Physical aspects of heterogeneities in multi-component lipid membranes. Adv. Colloid Interface Sci. 2014, 208, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, V.; Wan, C.; Tamm, L.K. Domain coupling in asymmetric lipid bilayers. Biochim. Biophys. Acta Biomembr. 2009, 1788, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickels, J.D.; Smith, J.C.; Cheng, X. Lateral organization, bilayer asymmetry, and inter-leaflet coupling of biological membranes. Chem. Phys. Lipids 2015, 192, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Marquardt, D.; Geier, B.; Pabst, G. Asymmetric lipid membranes: Towards more realistic model systems. Membranes 2015, 5, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Risselada, H.J.; Marrink, S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA 2008, 105, 17367–17372. [Google Scholar] [CrossRef] [Green Version]
- Garg, S.; Rühe, J.; Lüdtke, K.; Jordan, R.; Naumann, C.A. Domain registration in raft-mimicking lipid mixtures studied using polymer-tethered lipid bilayers. Biophys. J. 2007, 92, 1263–1270. [Google Scholar] [CrossRef]
- Collins, M.D. Interleaflet coupling mechanisms in bilayers of lipids and cholesterol. Biophys. J. 2008, 94, L32–L34. [Google Scholar] [CrossRef]
- May, S. Trans-monolayer coupling of fluid domains in lipid bilayers. Soft Matter 2009, 5, 3148–3156. [Google Scholar] [CrossRef]
- Williamson, J.J.; Olmsted, P.D. Registered and antiregistered phase separation of mixed amphiphilic bilayers. Biophys. J. 2015, 108, 1963–1976. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.D.; Feigenson, G.W. Presence and role of midplane cholesterol in lipid bilayers containing registered or antiregistered phase domains. J. Phys. Chem. B 2018, 122, 8193–8200. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.J.; Loew, S.; May, S. Influence of monolayer-monolayer coupling on the phase behavior of a fluid lipid bilayer. Biophys. J. 2007, 93, 4268–4277. [Google Scholar] [CrossRef] [PubMed]
- Putzel, G.G.; Schick, M. Phase behavior of a model bilayer membrane with coupled leaves. Biophys. J. 2008, 94, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Putzel, G.G.; Uline, M.J.; Szleifer, I.; Schick, M. Interleaflet coupling and domain registry in phase-separated lipid bilayers. Biophys. J. 2011, 100, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Funkhouser, C.M.; Mayer, M.; Solis, F.J.; Thornton, K. Effects of interleaflet coupling on the morphologies of multicomponent lipid bilayer membranes. J. Chem. Phys. 2013, 138, 024909. [Google Scholar] [CrossRef]
- Williamson, J.J.; Olmsted, P.D. Kinetics of symmetry and asymmetry in a phase-separating bilayer membrane. Phys. Rev. E 2015, 92, 052721. [Google Scholar] [CrossRef] [Green Version]
- Fowler, P.W.; Williamson, J.J.; Sansom, M.S.; Olmsted, P.D. Roles of interleaflet coupling and hydrophobic mismatch in lipid membrane phase-separation kinetics. J. Am. Chem. Soc. 2016, 138, 11633–11642. [Google Scholar] [CrossRef]
- Harder, T. Formation of functional cell membrane domains: The interplay of lipid–and protein–mediated interactions. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2003, 358, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M. Do proteins facilitate the formation of cholesterol-rich domains? Biochim. Biophys. Acta Biomembr. 2004, 1666, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epand, R.M. Proteins and cholesterol-rich domains. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1576–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulacu, M.; Peériole, X.; Marrink, S.J. In silico design of robust bolalipid membranes. Biomacromolecules 2011, 13, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Killian, J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta Biomembr. 1998, 1376, 401–416. [Google Scholar] [CrossRef]
- Ackerman, D.G.; Feigenson, G.W. Multiscale modeling of four-component lipid mixtures: Domain composition, size, alignment, and properties of the phase interface. J. Phys. Chem. B 2015, 119, 4240–4250. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, K.; Brownholland, D.; Carnini, A.; Thompson, D.H.; Johnston, L.J. AFM investigations of phase separation in supported membranes of binary mixtures of POPC and an eicosanyl-based bisphosphocholine bolalipid. Langmuir 2010, 26, 8525–8533. [Google Scholar] [CrossRef]
- Sun, H.; Chen, L.; Gao, L.; Fang, W. Nanodomain formation of ganglioside GM1 in lipid membrane: Effects of cholera toxin-mediated cross-linking. Langmuir 2015, 31, 9105–9114. [Google Scholar] [CrossRef]
- Safran, S. Statistical Thermodynamics of Surfaces, Interfaces, and Membranes; Perseus Books Group: New York, NY, USA, 1994. [Google Scholar]
- Davies, H.T. Statistical Mechanics of Phases, Interfaces, and Thin Films; VCH Publishers: New York, NY, USA, 1996. [Google Scholar]
- Sternberg, B.; L’Hostis, C.; Whiteway, C.A.; Watts, A. The essential role of specific Halobacterium halobium polar lipids in 2D-array formation of bacteriorhodopsin. Biochim. Biophys. Acta Biomembr. 1992, 1108, 21–30. [Google Scholar] [CrossRef]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I.; Barceló-Coblijn, G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008, 12, 829–875. [Google Scholar] [CrossRef]
- Heidemann, R.A.; Khalil, A.M. The calculation of critical points. AIChE J. 1980, 26, 769–779. [Google Scholar] [CrossRef]
- Akasaka, R. Calculation of the critical point for mixtures using mixture models based on Helmholtz energy equations of state. Fluid Phase Equilib. 2008, 263, 102–108. [Google Scholar] [CrossRef]
- Bell, I.H.; Jäger, A. Calculation of critical points from Helmholtz-energy-explicit mixture models. Fluid Phase Equilib. 2017, 433, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Fischer, T.; Risselada, H.J.; Vink, R.L. Membrane lateral structure: The influence of immobilized particles on domain size. Phys. Chem. Chem. Phys. 2012, 14, 14500–14508. [Google Scholar] [CrossRef] [PubMed]
- Schmid, F. Physical mechanisms of micro-and nanodomain formation in multicomponent lipid membranes. Biochim. Biophys. Acta Biomembr. 2017, 1859, 509–528. [Google Scholar] [CrossRef] [PubMed]
- Rosetti, C.M.; Mangiarotti, A.; Wilke, N. Sizes of lipid domains: What do we know from artificial lipid membranes? What are the possible shared features with membrane rafts in cells? Biochim. Biophys. Acta Biomembr. 2017, 1859, 789–802. [Google Scholar] [CrossRef]
- Sadeghi, S.; Müller, M.; Vink, R.L. Raft formation in lipid bilayers coupled to curvature. Biophys. J. 2014, 107, 1591–1600. [Google Scholar] [CrossRef]
- May, S.; Harries, D.; Ben-Shaul, A. Macroion-induced compositional instability of binary fluid membranes. Phys. Rev. Lett. 2002, 89, 268102. [Google Scholar] [CrossRef]
- Palmieri, B.; Safran, S.A. Hybrid lipids increase the probability of fluctuating nanodomains in mixed membranes. Langmuir 2013, 29, 5246–5261. [Google Scholar] [CrossRef]
- Ngamsaad, W.; May, S.; Wagner, A.J.; Triampo, W. Pinning of domains for fluid–fluid phase separation in lipid bilayers with asymmetric dynamics. Soft Matter 2011, 7, 2848–2857. [Google Scholar] [CrossRef]
- Witkowski, T.; Backofen, R.; Voigt, A. The influence of membrane bound proteins on phase separation and coarsening in cell membranes. Phys. Chem. Chem. Phys. 2012, 14, 14509–14515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honigmann, A.; Sadeghi, S.; Keller, J.; Hell, S.W.; Eggeling, C.; Vink, R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife 2014, 3, e01671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossa, G.V.; Roth, J.; May, S. Modeling lipid–lipid correlations across a bilayer membrane using the quasi-chemical approximation. Langmuir 2015, 31, 9924–9932. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, A.; Anikeev, V. Calculation of spinodal line and critical point of a mixture. Theor. Found. Chem. Eng. 2000, 34, 51–58. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bossa, G.V.; Gunderson, S.; Downing, R.; May, S. Role of Transmembrane Proteins for Phase Separation and Domain Registration in Asymmetric Lipid Bilayers. Biomolecules 2019, 9, 303. https://doi.org/10.3390/biom9080303
Bossa GV, Gunderson S, Downing R, May S. Role of Transmembrane Proteins for Phase Separation and Domain Registration in Asymmetric Lipid Bilayers. Biomolecules. 2019; 9(8):303. https://doi.org/10.3390/biom9080303
Chicago/Turabian StyleBossa, Guilherme Volpe, Sean Gunderson, Rachel Downing, and Sylvio May. 2019. "Role of Transmembrane Proteins for Phase Separation and Domain Registration in Asymmetric Lipid Bilayers" Biomolecules 9, no. 8: 303. https://doi.org/10.3390/biom9080303
APA StyleBossa, G. V., Gunderson, S., Downing, R., & May, S. (2019). Role of Transmembrane Proteins for Phase Separation and Domain Registration in Asymmetric Lipid Bilayers. Biomolecules, 9(8), 303. https://doi.org/10.3390/biom9080303




