Post-Translational Modifications in NETosis and NETs-Mediated Diseases
Abstract
1. Introduction
2. Neutrophils in Health and Disease
2.1. Neutrophils
2.2. Antimicrobial Functions of Neutrophils
3. Neutrophil Death
3.1. Neutrophil Apoptosis
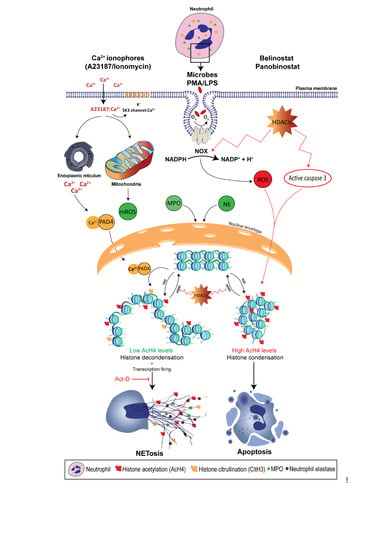
3.2. Neutrophil Extracellular Trap Formation (NETosis)
3.3. NOX-Dependent NETosis
3.4. Other Types of NETosis
4. Histone Modification in Neutrophils
4.1. Histone Citrullination
4.2. Histone Methylation
4.3. Histone Acetylation
4.4. Role of Post-Translational Modifications in NET-Mediated Diseases
4.5. Sepsis
4.6. Systemic Lupus Erythematosus (SLE)
4.7. Cancer
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mayadas, T.N.; Cullere, X.; Lowell, C.A. The Multifaceted Functions of Neutrophils. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 181–218. [Google Scholar] [CrossRef] [PubMed]
- Downey, D.G.; Bell, S.C.; Elborn, J.S. Neutrophils in cystic fibrosis. Thorax 2009, 64, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Kronbichler, A.; Park, D.D.Y.; Park, Y.M.; Moon, H.; Kim, H.; Choi, J.H.; Choi, Y.S.; Shim, S.; Lyu, I.S.; et al. Neutrophil extracellular traps (NETs) in autoimmune diseases: A comprehensive review. Autoimmun. Rev. 2017, 16, 1160–1173. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Stojkov, D.; Germic, N.; Simon, D.; Wang, X.; Benarafa, C.; Simon, H.U. Untangling “NETosis” from NETs. Eur. J. Immunol. 2019, 49, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Sollberger, G.; Tilley, D.O.; Zychlinsky, A. Neutrophil Extracellular Traps: The Biology of Chromatin Externalization. Dev. Cell 2018, 44, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Hamam, H.J.; Khan, M.A.; Palaniyar, N. Histone Acetylation Promotes Neutrophil Extracellular Trap Formation. Biomolecules 2019, 9, 32. [Google Scholar] [CrossRef]
- Hamam, H.J.; Palaniyar, N. Histone Deacetylase Inhibitors Dose-Dependently Switch Neutrophil Death from NETosis to Apoptosis. Biomolecules 2019, 9, 184. [Google Scholar] [CrossRef]
- Khan, M.A.; Palaniyar, N. Transcriptional firing helps to drive NETosis. Sci. Rep. 2017, 7, 41749. [Google Scholar] [CrossRef]
- Douda, D.N.; Khan, M.A.; Grasemann, H.; Palaniyar, N. SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc. Natl. Acad. Sci. USA 2015, 112, 2817–2822. [Google Scholar] [CrossRef]
- Yuen, J.; Pluthero, F.G.; Douda, D.N.; Riedl, M.; Cherry, A.; Ulanova, M.; Kahr, W.H.A.; Palaniyar, N.; Licht, C. NETosing neutrophils activate complement both on their own NETs and bacteria via alternative and non-alternative pathways. Front. Immunol. 2016, 7, 137. [Google Scholar] [CrossRef]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, L.; Yu, C.; Yu, D.; Yu, G. Histone Acetylation Modifiers in the Pathogenesis of Alzheimer’s Disease. Front. Cell. Neurosci. 2015, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Grigoryev, S.A.; Bulynko, Y.A.; Popova, E.Y. The end adjusts the means: Heterochromatin remodeling during terminal cell differentiation. Chromosom. Res. 2006, 14, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Kosak, S.T.; Groudine, M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004, 18, 1371–1384. [Google Scholar] [CrossRef]
- Lukášová, E.; Kořistek, Z.; Falk, M.; Kozubek, S.; Grigoryev, S.; Kozubek, M.; Ondřej, V.; Kroupová, I. Methylation of histones in myeloid leukemias as a potential marker of granulocyte abnormalities. J. Leukoc. Biol. 2005, 77, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Urdinguio, R.G.; Lopez, V.; Bayón, G.F.; Diaz de la Guardia, R.; Sierra, M.I.; García-Toraño, E.; Perez, R.F.; García, M.G.; Carella, A.; Pruneda, P.C.; et al. Chromatin regulation by Histone H4 acetylation at Lysine 16 during cell death and differentiation in the myeloid compartment. Nucleic Acids Res. 2019, 47, 5016–5037. [Google Scholar] [CrossRef] [PubMed]
- Lukášová, E.; Kořistek, Z.; Klabusay, M.; Ondřej, V.; Grigoryev, S.; Bačíková, A.; Řezáčová, M.; Falk, M.; Vávrová, J.; Kohútová, V.; et al. Granulocyte maturation determines ability to release chromatin NETs and loss of DNA damage response; these properties are absent in immature AML granulocytes. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 767–779. [Google Scholar] [CrossRef]
- Borregaard, N. Neutrophils, from Marrow to Microbes. Immunity 2010, 33, 657–670. [Google Scholar] [CrossRef]
- Edwards, S.W. The development and structure of mature neutrophils. In Biochemistry and Physiology of the Neutrophil; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar]
- Basu, S.; Hodgson, G.; Katz, M.; Dunn, A.R. Evaluation of role of G-CSF in the production, survival, and release of neutrophils from bone marrow into circulation. Blood 2002, 100, 854–861. [Google Scholar] [CrossRef]
- Galli, S.J.; Borregaard, N.; Wynn, T.A. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat. Immunol. 2011, 12, 1035. [Google Scholar] [CrossRef]
- Pillay, J.; Den Braber, I.; Vrisekoop, N.; Kwast, L.M.; De Boer, R.J.; Borghans, J.A.M.; Tesselaar, K.; Koenderman, L. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood 2010, 116, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Summers, C.; Rankin, S.M.; Condliffe, A.M.; Singh, N.; Peters, A.M.; Chilvers, E.R. Neutrophil kinetics in health and disease. Trends Immunol. 2010, 31, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Colotta, F.; Re, F.; Polentarutti, N.; Sozzani, S.; Mantovani, A. Modulation of granulocyte survival and programmed cell death by cytokines and bacterial products. Blood 1992, 80, 2012–2020. [Google Scholar] [PubMed]
- Wang, J.; Hossain, M.; Thanabalasuriar, A.; Gunzer, M.; Meininger, C.; Kubes, P. Visualizing the function and fate of neutrophils in sterile injury and repair. Science 2017, 358, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.M.; Everhart, J.E.; Byrd-Holt, D.D.; Tisdale, J.F.; Rodgers, G.P. Prevalence of neutropenia in the U.S. population: Age, sex, smoking status, and ethnic differences. Ann. Intern. Med. 2007, 146, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Biron, B.M.; Chung, C.-S.; Chen, Y.; Wilson, Z.; Fallon, E.A.; Reichner, J.S.; Ayala, A. PADI4 Deficiency Leads to Decreased Organ Dysfunction and Improved Survival in a Dual Insult Model of Hemorrhagic Shock and Sepsis. J. Immunol. 2018, 200, 1817–1828. [Google Scholar] [PubMed]
- Teng, T.-S.; Ji, A.; Ji, X.-Y.; Li, Y.-Z. Neutrophils and Immunity: From Bactericidal Action to Being Conquered. J. Immunol. Res. 2017, 2017. [Google Scholar] [CrossRef]
- Rigby, K.M.; DeLeo, F.R. Neutrophils in innate host defense against Staphylococcus aureus infections. In Seminars in Immunopathology; Springer: Berlin, Germany, 2012; pp. 237–259. [Google Scholar]
- Voisin, M.B.; Nourshargh, S. Neutrophil transmigration: Emergence of an adhesive cascade within venular walls. J. Innate Immun. 2013, 5, 336–347. [Google Scholar] [CrossRef]
- Sahakian, E.; Chen, J.; Powers, J.J.; Chen, X.; Maharaj, K.; Deng, S.L.; Achille, A.N.; Lienlaf, M.; Wang, H.W.; Cheng, F.; et al. Essential role for histone deacetylase 11 (HDAC11) in neutrophil biology. J. Leukoc. Biol. 2017, 102, 475–486. [Google Scholar] [CrossRef]
- Lee, W.L.; Harrison, R.E.; Grinstein, S. Phagocytosis by neutrophils. Microbes Infect. 2003, 5, 1299–1306. [Google Scholar] [CrossRef]
- Cheng, O.Z.; Palaniyar, N. NET balancing: A problem in inflammatory lung diseases. Front. Immunol. 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Cowland, J.B.; Borregaard, N. Granulopoiesis and granules of human neutrophils. Immunol. Rev. 2016, 273, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Roos, D.; Van Bruggen, R.; Meischl, C. Oxidative killing of microbes by neutrophils. Microbes Infect. 2003, 5, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Sengeløv, H.; Follin, P.; Kjeldsen, L.; Lollike, K.; Dahlgren, C.; Borregaard, N. Mobilization of granules and secretory vesicles during in vivo exudation of human neutrophils. J. Immunol. 1995, 154, 4157–4165. [Google Scholar] [PubMed]
- Masson, P.L. Lactoferrin, an iron-binbing protein Ni neutrophilic leukocytes. J. Exp. Med. 1969, 130, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.G.; Ganz, T.; Kinkade, J.M.; Selsted, M.E.; Lehrer, R.I.; Parmley, R.T. Defensin-rich dense granules of human neutrophils. Blood 1987, 70, 757–765. [Google Scholar] [PubMed]
- Campbell, E.J.; Campbell, M.A.; Owen, C.A. Bioactive Proteinase 3 on the Cell Surface of Human Neutrophils: Quantification, Catalytic Activity, and Susceptibility to Inhibition. J. Immunol. 2000, 165, 3366–3374. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.J.; Peppin, G.; Ortiz, X.; Ragsdale, C.; Test, S.T. Oxidative autoactivation of latent collagenase by human neutrophils. Science 1985, 227, 747–749. [Google Scholar] [CrossRef]
- Kasama, T.; Miwa, Y.; Isozaki, T.; Odai, T.; Adachi, M.; Kunkel, S. Neutrophil-Derived Cytokines: Potential Therapeutic Targets in Inflammation. Curr. Drug Targets Inflamm. Allergy 2005, 4, 273–279. [Google Scholar] [CrossRef]
- Kato, T.; Kitagawa, S. Regulation of Neutrophil Functions by Proinflammatory Cytokines. Int. J. Hematol. 2006, 84, 205–209. [Google Scholar] [CrossRef]
- McCracken, J.M.; Allen, L.A.H. Regulation of human neutrophil apoptosis and lifespan in health and disease. J. Cell Death 2014, 7, JCD-S11038. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.D.; Deleo, F.R. Neutrophil apoptosis and the resolution of infection. Immunol. Res. 2009, 43, 25–61. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Duckett, C.S. IAP proteins: Blocking the road to death’s door. Nat. Rev. Mol. Cell Biol. 2002, 3, 401. [Google Scholar] [CrossRef] [PubMed]
- Squier, M.K.T.; Sehnert, A.J.; Sellins, K.S.; Malkinson, A.M.; Takano, E.; Cohen, J.J. Calpain and calpastatin regulate neutrophil apoptosis. J. Cell. Physiol. 1999, 178, 311–319. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. The BCL2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2002, 2, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Milot, E.; Filep, J.G. Regulation of Neutrophil Survival/Apoptosis by Mcl-1. Sci. World J. 2011, 11, 1948–1962. [Google Scholar] [CrossRef]
- Andina, N.; Conus, S.; Schneider, E.M.; Fey, M.F.; Simon, H.U. Induction of Bim limits cytokine-mediated prolonged survival of neutrophils. Cell Death Differ. 2009, 16, 1248. [Google Scholar] [CrossRef]
- Villunger, A.; Scott, C.; Bouillet, P.; Strasser, A. Essential role for the BH3-only protein Bim but redundant roles for Bax, Bcl-2, and Bcl-w in the control of granulocyte survival. Blood 2003, 101, 2393–2400. [Google Scholar] [CrossRef]
- Murphy, B.M.; O’Neill, A.J.; Adrain, C.; Watson, R.W.G.; Martin, S.J. The Apoptosome Pathway to Caspase Activation in Primary Human Neutrophils Exhibits Dramatically Reduced Requirements for Cytochrome c. J. Exp. Med. 2003, 197, 625–632. [Google Scholar] [CrossRef]
- Altznauer, F.; Martinelli, S.; Yousefi, S.; Thürig, C.; Schmid, I.; Conway, E.M.; Schöni, M.H.; Vogt, P.; Mueller, C.; Fey, M.F.; et al. Inflammation-associated Cell Cycle–independent Block of Apoptosis by Survivin in Terminally Differentiated Neutrophils. J. Exp. Med. 2004, 199, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, P.; Gaehtgens, P.; Walzog, B. Bcl-Xl- and Bax-alpha-mediated regulation of apoptosis of human neutrophils via caspase-3. Blood 1999, 93, 3106–3115. [Google Scholar] [PubMed]
- Santos-Beneit, A.M.; Mollinedo, F. Expression of genes involved in initiation, regulation, and execution of apoptosis in human neutrophils and during neutrophil differentiation of HL-60 cells. J. Leukoc. Biol. 2000, 67, 712–724. [Google Scholar] [CrossRef] [PubMed]
- Moulding, D.A.; Akgul, C.; Derouet, M.; White, M.R.; Edwards, S.W. BCL-2 family expression in human neutrophils during delayed and accelerated apoptosis. J. Leukoc. Biol. 2001, 70, 783–792. [Google Scholar] [PubMed]
- Kasahara, Y.; Iwai, K.; Yachie, A.; Ohta, K.; Konno, A.; Seki, H.; Miyawaki, T.; Taniguchi, N. Involvement of reactive oxygen intermediates in spontaneous and CD95 (Fas/APO-1)-mediated apoptosis of neutrophils. Blood 1997, 89, 1748–1753. [Google Scholar]
- Aoshiba, K.; Yasui, S.; Hayashi, M.; Tamaoki, J.; Nagai, A. Role of p38-mitogen-activated protein kinase in spontaneous apoptosis of human neutrophils. J. Immunol. 1999, 162, 1692–1700. [Google Scholar] [PubMed]
- Zhang, B.; Hirahashi, J.; Cullere, X.; Mayadas, T.N. Elucidation of molecular events leading to neutrophil apoptosis following phagocytosis. Cross-talk between caspase 8, reactive oxygen species, and MAPK/ERK activation. J. Biol. Chem. 2003, 278, 28443–28454. [Google Scholar] [CrossRef] [PubMed]
- Takei, H.; Araki, A.; Watanabe, H.; Ichinose, A.; Sendo, F. Rapid killing of human neutrophils by the potent activator phorbol 12-myristate 13-acetate (PMA) accompanied by changes different from typical apoptosis or necrosis. J. Leukoc. Biol. 1996, 59, 229–240. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Von Köckritz-Blickwede, M.; Goldmann, O.; Thulin, P.; Heinemann, K.; Norrby-Teglund, A.; Rohde, M.; Medina, E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008, 111, 3070–3080. [Google Scholar] [CrossRef]
- Von Köckritz-Blickwede, M.; Chow, O.A.; Nizet, V. Fetal calf serum contains heat-stable nucleases that degrade neutrophil extracellular traps. Blood 2009, 114, 5245–5246. [Google Scholar] [CrossRef] [PubMed]
- Yipp, B.G.; Kubes, P. NETosis: How vital is it? Blood 2013, 122, 2784–2794. [Google Scholar] [CrossRef] [PubMed]
- Berghe, T.V.; Linkermann, A.; Jouan-Lanhouet, S.; Walczak, H.; Vandenabeele, P. Regulated necrosis: The expanding network of non-apoptotic cell death pathways. Nat. Rev. Mol. Cell Biol. 2014, 15, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Iba, T.; Hashiguchi, N.; Nagaoka, I.; Tabe, Y.; Murai, M. Neutrophil cell death in response to infection and its relation to coagulation. J. Intensive Care 2013, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Remijsen, Q.; Berghe, T.V.; Wirawan, E.; Asselbergh, B.; Parthoens, E.; De Rycke, R.; Noppen, S.; Delforge, M.; Willems, J.; Vandenabeele, P. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011, 21, 290. [Google Scholar] [CrossRef]
- Azzouz, D.; Palaniyar, N. ApoNETosis: Discovery of a novel form of neutrophil death with concomitant apoptosis and NETosis. Cell Death Dis. 2018, 9, 839. [Google Scholar] [CrossRef]
- Burg, N.D.; Pillinger, M.H. The neutrophil: Function and regulation in innate and humoral immunity. Clin. Immunol. 2001, 99, 7–17. [Google Scholar] [CrossRef]
- Rada, B.; Leto, T. Oxidative innate immune defenses by Nox/Duox Family NADPH oxidases. Contrib. Microbiol. 2008, 15, 164–187. [Google Scholar]
- Brown, D.I.; Griendling, K.K. Nox proteins in signal transduction. Free Radic. Biol. Med. 2009, 47, 1239–1253. [Google Scholar] [CrossRef]
- Karlsson, A.; Nixon, J.B.; McPhail, L.C. Phorbol myristate acetate induces neutrophil NADPH-oxidase activity by two separate signal transduction pathways: Dependent or independent of phosphatidylinositol 3-kinase. J. Leukoc. Biol. 2000, 67, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Farahvash, A.; Douda, D.N.; Licht, J.-C.C.; Grasemann, H.; Sweezey, N.; Palaniyar, N. JNK Activation Turns on LPS-And Gram-Negative Bacteria-Induced NADPH Oxidase-Dependent Suicidal NETosis. Sci. Rep. 2017, 7, 3409. [Google Scholar] [CrossRef] [PubMed]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. Myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Nauseef, W.M. How human neutrophils kill and degrade microbes: An integrated view. Immunol. Rev. 2007, 219, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Philip, L.M.; Cheung, G.; Vadakepeedika, S.; Grasemann, H.; Sweezey, N.; Palaniyar, N. Regulating NETosis: Increasing pH Promotes NADPH Oxidase-Dependent NETosis. Front. Med. 2018, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.N.; Breda, L.C.D.; Khan, M.A.; de Almeida, S.R.; Câmara, N.O.S.; Sweezey, N.; Palaniyar, N. Alkaline pH promotes NADPH oxidase-independent neutrophil extracellular trap formation: A matter of mitochondrial reactive oxygen species generation and citrullination and cleavage of histone. Front. Immunol. 2018, 8, 1849. [Google Scholar] [CrossRef] [PubMed]
- Vorobjeva, N.V.; Pinegin, B.V. Neutrophil Extracellular Traps: Mechanisms of formation and role in health and disease. Biochemistry 2014, 79, 1286–1296. [Google Scholar] [CrossRef]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A Novel Mechanism of Rapid Nuclear Neutrophil Extracellular Trap Formation in Response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef]
- Roos, D.; Voetman, A.A.; Meerhof, L.J. Functional activity of enucleated human polymorphonuclear leukocytes. J. Cell Biol. 1983, 97, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Giaglis, S.; Hasler, P.; Hahn, S. Efficient neutrophil extracellular trap induction requires mobilization of both intracellular and extracellular calcium pools and is modulated by cyclosporine A. PLoS ONE 2014, 9, e97088. [Google Scholar] [CrossRef] [PubMed]
- Slaba, I.; Wang, J.; Kolaczkowska, E.; Mcdonald, B.; Lee, W.Y.; Kubes, P. Imaging the dynamic platelet-neutrophil response in sterile liver injury and repair in mice. Hepatology 2015, 62, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.; Kubes, P. Innate Immune Cell Trafficking and Function During Sterile Inflammation of the Liver. Gastroenterology 2016, 151, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Christoffersson, G.; Henriksnäs, J.; Johansson, L.; Rolny, C.; Ahlström, H.; Caballero-Corbalan, J.; Segersvärd, R.; Permert, J.; Korsgren, O.; Carlsson, P.O.; et al. Clinical and experimental pancreatic islet transplantation to striated muscle: Establishment of a vascular system similar to that in native islets. Diabetes 2010, 59, 2569–2578. [Google Scholar] [CrossRef]
- Azzouz, L.; Cherry, A.; Riedl, M.; Khan, M.; Pluthero, F.G.; Kahr, W.H.A.; Palaniyar, N.; Licht, C. Relative antibacterial functions of complement and NETs: NETs trap and complement effectively kills bacteria. Mol. Immunol. 2018, 97, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps: Is immunity the second function of chromatin? J. Cell Biol. 2012, 198, 773–783. [Google Scholar] [CrossRef]
- Bianchi, M.; Hakkim, A.; Brinkmann, V.; Siler, U.; Seger, R.A.; Zychlinsky, A.; Reichenbach, J. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood 2009, 114, 2619–2622. [Google Scholar] [CrossRef]
- Jones, J.E.; Causey, C.P.; Knuckley, B.; Slack-Noyes, J.L.; Thompson, P.R. Protein arginine deiminase 4 (PADI4 ): Current understanding and future therapeutic potential. Curr. Opin. Drug Discov. Dev. 2009, 12, 616. [Google Scholar]
- Neeli, I.; Khan, S.N.; Radic, M. Histone Deimination As a Response to Inflammatory Stimuli in Neutrophils. J. Immunol. 2014, 180, 1895–1902. [Google Scholar] [CrossRef]
- Luo, Y.; Arita, K.; Bhatia, M.; Knuckley, B.; Lee, Y.H.; Stallcup, M.R.; Sato, M.; Thompson, P.R. Inhibitors and inactivators of protein arginine deiminase 4: Functional and structural characterization. Biochemistry 2006, 45, 11727–11736. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, M.; Lindberg, M.R.; Kennett, M.J.; Xiong, N.; Wang, Y. PADI4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010, 207, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.L.; Ng, G.; Tablin, F. Lipopolysaccharide-induced neutrophil extracellular trap formation in canine neutrophils is dependent on histone H3 citrullination by peptidylarginine deiminase. Vet. Immunol. Immunopathol. 2017, 193, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Neeli, I.; Radic, M. Opposition between PKC isoforms regulates histone deimination and neutrophil extracellular chromatin release. Front. Immunol. 2013, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Kenny, E.F.; Herzig, A.; Krüger, R.; Muth, A.; Mondal, S.; Thompson, P.R.; Brinkmann, V.; von Bernuth, H.; Zychlinsky, A. Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 2017, 6, e24437. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Franz, H.; Jacobs, S.A.; Allis, C.D.; Khorasanizadeh, S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 2008, 283, 19626–19635. [Google Scholar] [CrossRef] [PubMed]
- Byvoet, P.; Shepherd, G.R.; Hardin, J.M.; Noland, B.J. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch. Biochem. Biophys. 1972, 148, 558–567. [Google Scholar] [CrossRef]
- Murray, K. The Occurrence of epsilon-N-Methyl Lysine in Histones. Biochemistry 1963, 3, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtier, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Wang, H.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Struhl, K.; Zhang, Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002, 12, 1052–1058. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Cloos, P.A.C.; Christensen, J.; Agger, K.; Maiolica, A.; Rappsilber, J.; Antal, T.; Hansen, K.H.; Helin, K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 2006, 442, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of Histone Lysine Trimethylation by the JMJD2 Family of Histone Demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef]
- Chang, B.; Chen, Y.; Zhao, Y.; Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 2007, 318, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Webby, C.J.; Wolf, A.; Gromak, N.; Dreger, M.; Kramer, H.; Kessler, B.; Nielsen, M.L.; Schmitz, C.; Butler, D.S.; Yates, J.R.; et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 2009, 325, 90–93. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Pieterse, E.; Hofstra, J.; Berden, J.; Herrmann, M.; Dieker, J.; van der Vlag, J. Acetylated histones contribute to the immunostimulatory potential of neutrophil extracellular traps in systemic lupus erythematosus. Clin. Exp. Immunol. 2015, 179, 68–74. [Google Scholar] [CrossRef]
- Liu, C.L.; Tangsombatvisit, S.; Rosenberg, J.M.; Mandelbaum, G.; Gillespie, E.C.; Gozani, O.P.; Alizadeh, A.A.; Utz, P.J. Specific post-translational histone modifications of neutrophil extracellular traps as immunogens and potential targets of lupus autoantibodies. Arthritis Res. Ther. 2012, 14, R25. [Google Scholar] [CrossRef]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Rajakumara, E.; Law, J.A.; Simanshu, D.K.; Voigt, P.; Johnson, L.M.; Reinberg, D.; Patel, D.J.; Jacobsen, S.E. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011, 25, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Hashimshony, T.; Zhang, J.; Keshet, I.; Bustin, M.; Cedar, H. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 2003, 34, 187. [Google Scholar] [CrossRef] [PubMed]
- Lande-Diner, L.; Zhang, J.; Ben-Porath, I.; Amariglio, N.; Keshet, I.; Hecht, M.; Azuara, V.; Fisher, A.G.; Rechavi, G.; Cedar, H. Role of DNA methylation in stable gene repression. J. Biol. Chem. 2007, 282, 12194–12200. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.H.; Pertin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PADI4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Raijmakers, R.; Zendman, A.J.W.; Egberts, W.V.; Vossenaar, E.R.; Raats, J.; Soede-Huijbregts, C.; Rutjes, F.P.J.T.; van Veelen, P.A.; Drijfhout, J.W.; Pruijn, G.J.M. Methylation of Arginine Residues Interferes with Citrullination by Peptidylarginine Deiminases in vitro. J. Mol. Biol. 2007, 367, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone deimination antagonizes arginine methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone acetyltransferases. Histone Acetyltransferases 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Hollands, A.; Corriden, R.; Gysler, G.; Dahesh, S.; Olson, J.; Ali, S.R.; Kunkel, M.T.; Lin, A.E.; Forli, S.; Newton, A.C.; et al. Natural product anacardic acid from cashew nut shells stimulates neutrophil extracellular trap production and bactericidal activity. J. Biol. Chem. 2016, 291, 13964–13973. [Google Scholar] [CrossRef]
- Pandey, D.; Chen, F.; Patel, A.; Wang, C.Y.; Dimitropoulou, C.; Patel, V.S.; Rudic, R.D.; Stepp, D.W.; Fulton, D.J. SUMO1 negatively regulates reactive oxygen species production from NADPH oxidases. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1634–1642. [Google Scholar] [CrossRef]
- Johnstone, R.W.; Licht, J.D. Histone deacetylase inhibitors in cancer therapy: Is transcription the primary target? Cancer Cell 2003, 4, 13–18. [Google Scholar] [CrossRef]
- Yang, X.-J.; Grégoire, S. Class II histone deacetylases: From sequence to function, regulation, and clinical implication. Mol. Cell. Biol. 2005, 25, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Medicinal Chemistry of Sirtuin Inhibitors. Curr. Med. Chem. 2011, 18, 1936–1946. [Google Scholar] [CrossRef] [PubMed]
- Kankaanranta, H.; Janka-Junttila, M.; Ilmarinen-Salo, P.; Ito, K.; Jalonen, U.; Ito, M.; Adcock, I.M.; Moilanen, E.; Zhang, X. Histone deacetylase inhibitors induce apoptosis in human eosinophils and neutrophils. J. Inflamm. 2010, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yu, C.-W.; Duan, J.; Luo, M.; Wang, K.; Tian, G.; Cui, Y.; Wu, K. HDA6 Directly Interacts with DNA Methyltransferase MET1 and Maintains Transposable Element Silencing in Arabidopsis. Plant Physiol. 2012, 158, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Vaissière, T.; Sawan, C.; Herceg, Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. Rev. Mutat. Res. 2008, 659, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Coonrod, S.A.; Kraus, W.L.; Jelinek, M.A.; Stallcup, M.R. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. USA 2005, 102, 3611–3616. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Thompson, P.R. Protein Arginine Methylation and Citrullination in Epigenetic Regulation. ACS Chem. Biol. 2016, 11, 654–668. [Google Scholar] [CrossRef]
- Denis, H.; Deplus, R.; Putmans, P.; Yamada, M.; Metivier, R.; Fuks, F. Functional Connection between Deimination and Deacetylation of Histones. Mol. Cell. Biol. 2009, 29, 4982–4993. [Google Scholar] [CrossRef]
- Dokmanovic, M.; Marks, P.A. Prospects: Histone deacetylase inhibitors. J. Cell. Biochem. 2005, 96, 293–304. [Google Scholar] [CrossRef]
- Marks, P.A.; Dokmanovic, M. Histone deacetylase inhibitors: Discovery and development as anticancer agents. Expert Opin. Investig. Drugs 2005, 14, 1497–1511. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, P.M.; Cole, K.E.; Dowling, D.P.; Christianson, D.W. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 2011, 21, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Eom, G.H. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med. J. 2016, 52, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-Cell Lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.A.; Xu, W.S. Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell. Biochem. 2009, 107, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.M. Belinostat: First global approval. Drugs 2014, 74, 1445–1453. [Google Scholar] [CrossRef]
- Molife, L.R.; de Bono, J.S. Belinostat: Clinical applications in solid tumors and lymphoma. Expert Opin. Investig. Drugs 2011, 20, 1723–1732. [Google Scholar] [CrossRef]
- Plumb, J.A.; Finn, P.W.; Williams, R.J.; Bandara, M.J.; Romero, M.R.; Watkins, C.J.; La Thangue, N.B.; Brown, R. Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol. Cancer Ther. 2003, 2, 721–728. [Google Scholar]
- Steele, N.L.; Plumb, J.A.; Vidal, L.; Tjørnelund, J.; Knoblauch, P.; Rasmussen, A.; Ooi, C.E.; Buhl-Jensen, P.; Brown, R.; Evans, T.R.J.; et al. A phase 1 pharmacokinetic and pharmacodynamic study of the histone deacetylase inhibitor belinostat in patients with advanced solid tumors. Clin. Cancer Res. 2008, 14, 804–810. [Google Scholar] [CrossRef]
- Tumber, A.; Collins, L.S.; Petersen, K.D.; Thougaard, A.; Christiansen, S.J.; Dejligbjerg, M.; Jensen, P.B.; Sehested, M.; Ritchie, J.W.A. The histone deacetylase inhibitor PXD101 synergises with 5-fluorouracil to inhibit colon cancer cell growth in vitro and in vivo. Cancer Chemother. Pharmacol. 2007, 60, 275–283. [Google Scholar] [CrossRef]
- Kong, L.R.; Tan, T.Z.; Ong, W.R.; Bi, C.; Huynh, H.; Lee, S.C.; Chng, W.J.; Eichhorn, P.J.A.; Goh, B.C. Belinostat exerts antitumor cytotoxicity through the ubiquitin-proteasome pathway in lung squamous cell carcinoma. Mol. Oncol. 2017, 11, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Valiuliene, G.; Stirblyte, I.; Cicenaite, D.; Kaupinis, A.; Valius, M.; Navakauskiene, R. Belinostat, a potent HDACi, exerts antileukaemic effect in human acute promyelocytic leukaemia cells via chromatin remodeling. J. Cell. Mol. Med. 2015, 19, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ara, G.; Mills, E.; LaRochelle, W.J.; Lichenstein, H.S.; Jeffers, M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int. J. Cancer 2008, 122, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Qian, X. Activity of PXD101, a histone deacetylase inhibitor, in preclinical ovarian cancer studies. Mol. Cancer Ther. 2006, 5, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Buckley, M.T.; Yoon, J.; Yee, H.; Chiriboga, L.; Liebes, L.; Ara, G.; Qian, X.; Bajorin, D.F.; Sun, T.-T.; Wu, X.-R.; et al. The histone deacetylase inhibitor belinostat (PXD101) suppresses bladder cancer cell growth in vitro and in vivo. J. Transl. Med. 2007, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Z.; Kwitkowski, V.E.; Del Valle, P.L.; Ricci, M.S.; Saber, H.; Habtemariam, B.A.; Bullock, J.; Bloomquist, E.; Shen, Y.L.; Chen, X.H.; et al. FDA approval: Belinostat for the treatment of patients with relapsed or refractory peripheral T-cell lymphoma. Clin. Cancer Res. 2015, 21, 2666–2670. [Google Scholar] [CrossRef]
- (FDA), U.S.F. & D. administration Press Announcements - FDA approves Farydak for treatment of multiple myeloma. Available online: https://www.cancer.org/latest-news/fda-approves-farydak-panobinostat-for-multiple-myeloma.html (accessed on 25 June 2019).
- Garnock-Jones, K.P. Panobinostat: First global approval. Drugs 2015, 75, 695–704. [Google Scholar] [CrossRef]
- Atadja, P. Development of the pan-DAC inhibitor panobinostat (LBH589): Successes and challenges. Cancer Lett. 2009, 280, 233–241. [Google Scholar] [CrossRef]
- Cheng, T.; Grasse, L.; Shah, J.; Chandra, J. Panobinostat, a pan-histone deacetylase inhibitor: Rationale for and application to treatment of multiple myeloma. Drugs Today 2015, 51, 491–504. [Google Scholar] [CrossRef]
- Moore, D. Panobinostat (Farydak): A Novel Option for the Treatment of Relapsed or Relapsed and Refractory Multiple Myeloma. Pharm. Ther. 2016, 41, 296. [Google Scholar]
- Cashen, A.; Juckett, M.; Jumonville, A.; Litzow, M.; Flynn, P.J.; Eckardt, J.; LaPlant, B.; Laumann, K.; Erlichman, C.; DiPersio, J. Phase II study of the histone deacetylase inhibitor belinostat (PXD101) for the treatment of myelodysplastic syndrome (MDS). Ann. Hematol. 2012, 91, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Dincman, T.A.; Beare, J.E.; Ohri, S.S.; Gallo, V.; Hetman, M.; Whittemore, S.R. Histone deacetylase inhibition is cytotoxic to oligodendrocyte precursor cells in vitro and in vivo. Int. J. Dev. Neurosci. 2016, 54, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Savickiene, J.; Treigyte, G.; Valiuliene, G.; Stirblyte, I.; Navakauskiene, R. Epigenetic and molecular mechanisms underlying the antileukemic activity of the histone deacetylase inhibitor belinostat in human acute promyelocytic leukemia cells. Anticancer Drugs 2014, 25, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.J.; Huang, K.K.; Yang, M.; Qiao, L.; Wang, Q.; Ye, J.Y.; Zhou, H.S.; Yi, Z.S.; Wu, F.Q.; Wang, Z.X.; et al. Synergistic effect of panobinostat and bortezomib on chemoresistant acute myelogenous leukemia cells via AKT and NF-κB pathways. Cancer Lett. 2012, 326, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Matthay, M.A.; Ware, L.B.; Zimmerman, G.A. The acute respiratory distress syndrome. J. Clin. Investig. 2012, 122, 2731–2740. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, X.; Pan, P.; Zhang, L.; Hu, Y.; Tan, H.; Wu, D.; Liu, B.; Li, H.; Li, H.; et al. Neutrophil extracellular traps are indirectly triggered by lipopolysaccharide and contribute to acute lung injury. Sci. Rep. 2016, 6, 37252. [Google Scholar] [CrossRef]
- Ota, C.; Yamada, M.; Fujino, N.; Motohashi, H.; Tando, Y.; Takei, Y.; Suzuki, T.; Takahashi, T.; Kamata, S.; Makiguchi, T.; et al. Histone deacetylase inhibitor restores surfactant protein-C expression in alveolar-epithelial type II cells and attenuates bleomycin-induced pulmonary fibrosis in vivo. Exp. Lung Res. 2015, 41, 422–434. [Google Scholar] [CrossRef]
- Sperling, C.; Fischer, M.; Maitz, M.F.; Werner, C. Neutrophil extracellular trap formation upon exposure of hydrophobic materials to human whole blood causes thrombogenic reactions. Biomater. Sci. 2017, 5, 1998–2008. [Google Scholar] [CrossRef]
- Martinod, K.; Demers, M.; Fuchs, T.A.; Wong, S.L.; Brill, A.; Gallant, M.; Hu, J.; Wang, Y.; Wagner, D.D. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 8674–8679. [Google Scholar] [CrossRef]
- Lipinska-Gediga, M. Neutrophils, NETs, NETosis—Old or new factors in sepsis and septic shock? Anestezjol. Intens. Ter. 2017, 49, 235–240. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Pelayo, R.; Monestier, M.; Ammollo, C.T.; Semeraro, F.; Taylor, F.B.; Esmon, N.L.; Lupu, F.; Esmon, C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009, 15, 1318. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.R.; Ma, A.C.; Tavener, S.A.; McDonald, B.; Goodarzi, Z.; Kelly, M.M.; Patel, K.D.; Chakrabarti, S.; McAvoy, E.; Sinclair, G.D.; et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Saffarzadeh, M.; Juenemann, C.; Queisser, M.A.; Lochnit, G.; Barreto, G.; Galuska, S.P.; Lohmeyer, J.; Preissner, K.T. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: A predominant role of histones. PLoS ONE 2012, 7, e32366. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, B.M.; Fisher, B.J.; Kraskauskas, D.; Farkas, D.; Brophy, D.F.; Fowler, A.A.; Natarajan, R. Vitamin C: A novel regulator of neutrophil extracellular trap formation. Nutrients 2013, 5, 3131–3150. [Google Scholar] [CrossRef] [PubMed]
- Claushuis, T.A.M.; van der Donk, L.E.H.; Luitse, A.L.; van Veen, H.A.; van der Wel, N.N.; van Vught, L.A.; Roelofs, J.J.T.H.; de Boer, O.J.; Lankelma, J.M.; Boon, L.; et al. Role of Peptidylarginine Deiminase 4 in Neutrophil Extracellular Trap Formation and Host Defense during Klebsiella pneumoniae—Induced Pneumonia-Derived Sepsis. J. Immunol. 2018, 201, 1241–1252. [Google Scholar] [CrossRef]
- Nomura, K.; Miyashita, T.; Yamamoto, Y.; Munesue, S.; Harashima, A.; Takayama, H.; Fushida, S.; Ohta, T. Citrullinated Histone H3: Early Biomarker of Neutrophil Extracellular Traps in Septic Liver Damage. J. Surg. Res. 2019, 234, 132–138. [Google Scholar] [CrossRef]
- Warford, J.; Lamport, A.C.; Kennedy, B.; Easton, A.S. Human Brain Chemokine and Cytokine Expression in Sepsis: A Report of Three Cases. Can. J. Neurol. Sci. 2017, 44, 96–104. [Google Scholar] [CrossRef]
- Alamdari, N.; Smith, I.J.; Aversa, Z.; Hasselgren, P.-O. Sepsis and glucocorticoids upregulate p300 and downregulate HDAC6 expression and activity in skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R509–R520. [Google Scholar] [CrossRef]
- Cheng, F.; Lienlaf, M.; Perez-Villarroel, P.; Wang, H.W.; Lee, C.; Woan, K.; Woods, D.; Knox, T.; Bergman, J.; Pinilla-Ibarz, J.; et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol. Immunol. 2014, 60, 44–53. [Google Scholar] [CrossRef]
- von Knethen, A.; Brüne, B. Histone Deacetylation Inhibitors as Therapy Concept in Sepsis. Int. J. Mol. Sci. 2019, 20, 346. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, F.; Woan, K.; Sahakian, E.; Merino, O.; Rock-Klotz, J.; Vicente-Suarez, I.; Pinilla-Ibarz, J.; Wright, K.L.; Seto, E.; et al. Histone Deacetylase Inhibitor LAQ824 Augments Inflammatory Responses in Macrophages through Transcriptional Regulation of IL-10. J. Immunol. 2011, 186, 3986–3996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jin, S.; Wang, C.; Jiang, R.; Wan, J. Histone deacetylase inhibitors attenuate acute lung injury during cecal ligation and puncture-induced polymicrobial sepsis. World J. Surg. 2010, 34, 1676–1683. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.H.; Li, G.M.; Jia, M.; Zhu, S.H.; Gao, D.P.; Fan, Y.X.; Wu, J.; Yang, J.J. Valproic acid attenuates lipopolysaccharide-induced acute lung injury in mice. Inflammation 2013, 36, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Cui, K.; Schones, D.E.; Barski, A.; Peng, W.; Zhao, K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009, 138, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Gibot, S.; Alauzet, C.; Massin, F.; Sennoune, N.; Faure, G.C.; Béné, M.; Lozniewski, A.; Bollaert, P.; Lévy, B. Modulation of the Triggering Receptor Expressed on Myeloid Cells—1 Pathway during Pneumonia in Rats. J. Infect. Dis. 2006, 194, 975–983. [Google Scholar] [CrossRef]
- Yuan, Z.; Syed, M.A.; Panchal, D.; Rogers, D.; Joo, M.; Sadikot, R.T. Curcumin mediated epigenetic modulation inhibits TREM-1 expression in response to lipopolysaccharide. Int. J. Biochem. Cell Biol. 2012, 44, 2032–2043. [Google Scholar] [CrossRef]
- Rahman, A.; Isenberg, D.A. Systemic Lupus Erythematosus. N. Engl. J. Med. 2008, 358, 929–939. [Google Scholar] [CrossRef]
- Hakkim, A.; Furnrohr, B.G.; Amann, K.; Laube, B.; Abed, U.A.; Brinkmann, V.; Herrmann, M.; Voll, R.E.; Zychlinsky, A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA 2010, 107, 9813–9818. [Google Scholar] [CrossRef]
- Leffler, J.; Martin, M.; Gullstrand, B.; Tyden, H.; Lood, C.; Truedsson, L.; Bengtsson, A.A.; Blom, A.M. Neutrophil Extracellular Traps That Are Not Degraded in Systemic Lupus Erythematosus Activate Complement Exacerbating the Disease. J. Immunol. 2012, 188, 3522–3531. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, N.; Upadhyay, J.; Neeli, I.; Khan, S.; Pattanaik, D.; Myers, L.; Kirou, K.A.; Hellmich, B.; Knuckley, B.; Thompson, P.R.; et al. Felty’s syndrome autoantibodies bind to deiminated histones and neutrophil extracellular chromatin traps. Arthritis Rheum. 2012, 64, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, N.; Neeli, I.; Schall, N.; Wan, H.; Desiderio, D.M.; Csernok, E.; Thompson, P.R.; Dali, H.; Briand, J.P.; Muller, S.; et al. Deimination of linker histones links neutrophil extracellular trap release with autoantibodies in systemic autoimmunity. FASEB J. 2014, 28, 2840–2851. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.S.; Zhao, W.; Luo, W.; Subramanian, V.; O’Dell, A.A.; Yalavarthi, S.; Hodgin, J.B.; Eitzman, D.T.; Thompson, P.R.; Kaplan, M.J. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J. Clin. Investig. 2013, 123, 2981–2993. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.S.; Subramanian, V.; O’Dell, A.A.; Yalavarthi, S.; Zhao, W.; Smith, C.K.; Hodgin, J.B.; Thompson, P.R.; Kaplan, M.J. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann. Rheum. Dis. 2015, 74, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Fiskus, W.; Ren, Y.; Mohapatra, A.; Bali, P.; Mandawat, A.; Rao, R.; Herger, B.; Yang, Y.; Atadja, P.; Wu, J.; et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-α levels and transcriptional activity: A result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin. Cancer Res. 2007, 13, 4882–4890. [Google Scholar] [CrossRef] [PubMed]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W. Histone-deacetylase inhibitors: Novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 2002, 1, 287–299. [Google Scholar] [CrossRef]
- Park, J.H.; Jung, Y.; Kim, T.Y.; Kim, S.G.; Jong, H.S.; Lee, J.W.; Kim, D.K.; Lee, J.S.; Kim, N.K.; Kim, T.Y.; et al. Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation. Clin. Cancer Res. 2004, 10, 5271–5281. [Google Scholar] [CrossRef]
- Huang, B.H.; Laban, M.; Leung, C.H.-W.; Lee, L.; Lee, C.K.; Salto-Tellez, M.; Raju, G.C.; Hooi, S.C. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005, 12, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Swierczak, A.; Mouchemore, K.A.; Hamilton, J.A.; Anderson, R.L. Neutrophils: Important contributors to tumor progression and metastasis. Cancer Metastasis Rev. 2015, 34, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Demers, M.; Wong, S.L.; Martinod, K.; Gallant, M.; Cabral, J.E.; Wang, Y.; Wagner, D.D. Priming of neutrophils toward NETosis promotes tumor growth. Oncoimmunology 2016, 5, e1134073. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, E.; Rother, N.; Garsen, M.; Hofstra, J.M.; Satchell, S.C.; Hoffmann, M.; Loeven, M.A.; Knaapen, H.K.; Van Der Heijden, O.W.H.; Berden, J.H.M.; et al. Neutrophil Extracellular Traps Drive Endothelial-to-Mesenchymal Transition. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. 2013, 123, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
- Thålin, C.; Lundström, S.; Seignez, C.; Daleskog, M.; Lundström, A.; Henriksson, P.; Helleday, T.; Phillipson, M.; Wallén, H.; Demers, M. Citrullinated histone H3 as a novel prognostic blood marker in patients with advanced cancer. PLoS ONE 2018, 13, e0191231. [Google Scholar] [CrossRef] [PubMed]
- Sen, G.S.; Mohanty, S.; Hossain, D.M.S.; Bhattacharyya, S.; Banerjee, S.; Chakraborty, J.; Saha, S.; Ray, P.; Bhattacharjee, P.; Mandal, D.; et al. Curcumin enhances the efficacy of chemotherapy by tailoring p65NFκB-p300 cross-talk in favor of p53-p300 in breast cancer. J. Biol. Chem. 2011, 286, 42232–42247. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Zhu, D.; Xue, Z.; Mao, H. Alteration of histone H3 lysine 9 dimethylation in peripheral white blood cells of septic patients with trauma and cancer. Mol. Med. Rep. 2016, 14, 5467–5474. [Google Scholar] [CrossRef]
- Simiele, F.; Recchiuti, A.; Patruno, S.; Plebani, R.; Pierdomenico, A.M.; Codagnone, M.; Romano, M. Epigenetic regulation of the formyl peptide receptor 2 gene. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 1252–1258. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.B.; Chen, L.Y.; Huang, L.; Dong, R.Z. Belinostat-induced apoptosis and growth inhibition in pancreatic cancer cells involve activation of TAK1-AMPK signaling axis. Biochem. Biophys. Res. Commun. 2013, 437, 1–6. [Google Scholar] [CrossRef]
- Lin, S.F.; Lin, J.D.; Chou, T.C.; Huang, Y.Y.; Wong, R.J. Utility of a Histone Deacetylase Inhibitor (PXD101) for Thyroid Cancer Treatment. PLoS ONE 2013, 8, e77684. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamam, H.J.; Palaniyar, N. Post-Translational Modifications in NETosis and NETs-Mediated Diseases. Biomolecules 2019, 9, 369. https://doi.org/10.3390/biom9080369
Hamam HJ, Palaniyar N. Post-Translational Modifications in NETosis and NETs-Mediated Diseases. Biomolecules. 2019; 9(8):369. https://doi.org/10.3390/biom9080369
Chicago/Turabian StyleHamam, Hussein J., and Nades Palaniyar. 2019. "Post-Translational Modifications in NETosis and NETs-Mediated Diseases" Biomolecules 9, no. 8: 369. https://doi.org/10.3390/biom9080369
APA StyleHamam, H. J., & Palaniyar, N. (2019). Post-Translational Modifications in NETosis and NETs-Mediated Diseases. Biomolecules, 9(8), 369. https://doi.org/10.3390/biom9080369