Gene and Metabolite Integration Analysis through Transcriptome and Metabolome Brings New Insight into Heat Stress Tolerance in Potato (Solanum tuberosum L.)
Abstract
:1. Introduction
2. Results
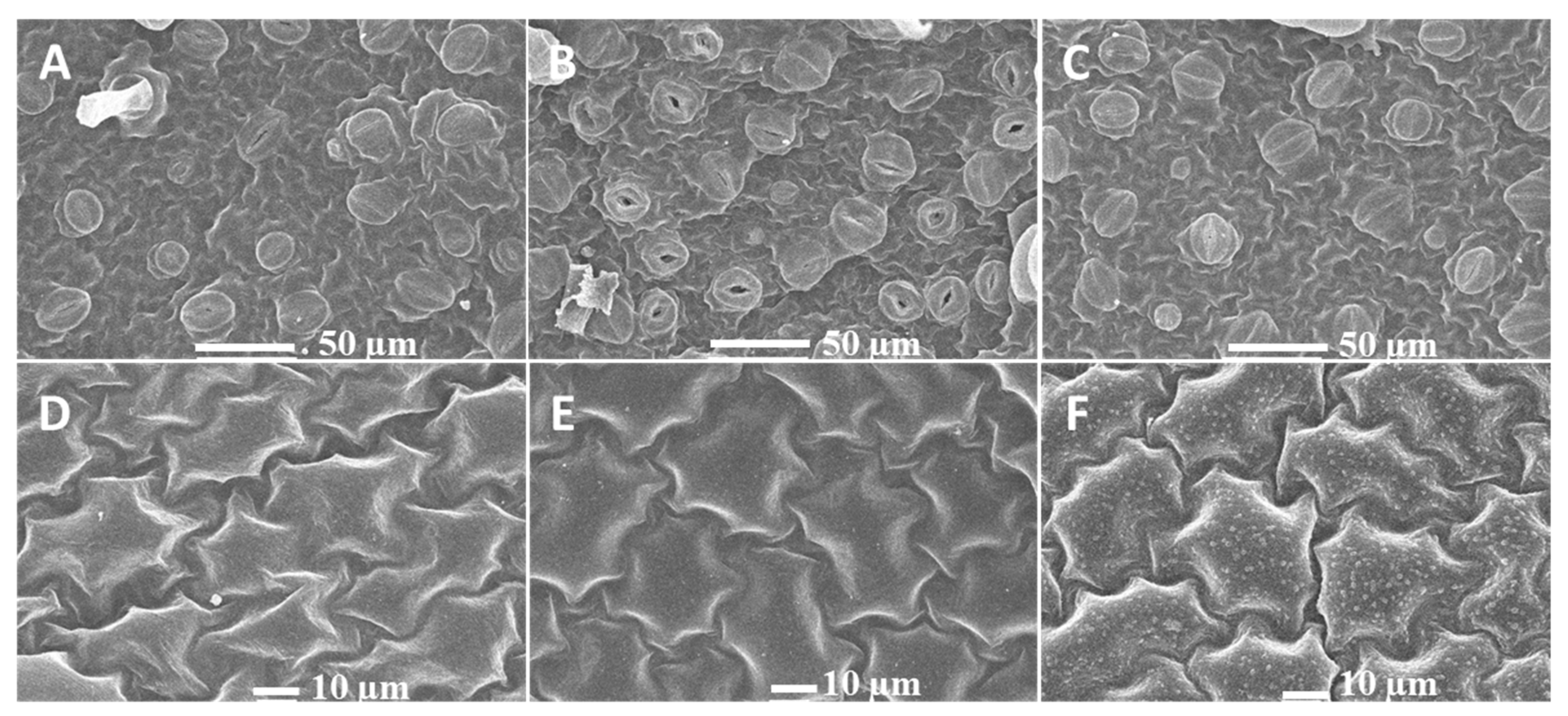
2.1. Phenotypic Responses to Heat Stress in Potato
2.2. Heat Stress Induced Transcriptional Changes in Potato Leaf
2.3. Functional Category Enrichment
2.4. Transcriptional Regulation Associated with Short and Prolonged Heat Stress
2.5. Comparison of Heat Stress-Responsive in Potato with Heat Stress-Related Genes in Arabidopsis thaliana Shoots and Drought Stress-Related Genes in Potato
2.6. Transient Expression of Heat Induced Genes in Nicotiana benthamiana Leaves
2.7. RNA-seq Validation by RT–qPCR
2.8. Short and Prolonged Heat Stress Cause Metabolic Alterations in Potato Leaf
3. Discussion
3.1. Short Heat Stress Maintaining the Stomata Open and Continued Stress Affect Photosynthetic Parameters
3.2. Upregulated Protective Proteins Are Characterized in Response to Heat Stress
3.3. General and Specific Responses to Heat-Stress in Potato
3.4. Secondary Metabolism and Amino Acid Metabolism Involved in Heat Stress
4. Materials and Methods
4.1. Plant Material
4.2. Physiological Indices and Metabolic Assays
4.3. RNA Samples Preparation and High-Throughput Sequencing
4.4. Metabolite Extraction, Measurement and Analysis
4.5. Transient Expression in Nicotiana benthamiana
4.6. Real-Time qRT–PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Panel on Climate Change. Summary for policymakers. In Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation; Field, C.B., Barros, V., Stocker, T.F., Qin, D.H., Dokken, D.J., Ebi, K.L., Mastrandrea, M.D., Mach, K.J., Plattner, G.-K., Allen, S.K., Eds.; Cambridge University Press: Cambridge, UK, 2012; p. 13. [Google Scholar]
- Tang, R.; Niu, S.; Zhang, G.; Chen, G.; Haroon, M.; Yang, Q.; Rajora, O.P.; Li, X.-Q. Physiological and growth responses of potato cultivars to heat stress. Botany 2018, 96, 897–912. [Google Scholar] [CrossRef]
- Rosenzweig, C.; Elliott, J.; Deryng, D.; Ruane, A.C.; Müller, C.; Arneth, A.; Boote, K.J.; Folberth, C.; Glotter, M.; Khabarov, N.; et al. Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc. Natl. Acad. Sci. USA 2014, 111, 3268–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rykaczewska, K. The impact of high temperature during growing season on potato cultivars with different response to environmental stresses. Am. J. Potato Res. 2013, 4, 2386–2393. [Google Scholar] [CrossRef] [Green Version]
- Levy, D.; Veilleux, R.E. Adaptation of potato to high temperatures and salinity-a review. Am. J. Potato Res. 2007, 84, 487–506. [Google Scholar] [CrossRef]
- Hancock, R.D.; Morris, W.L.; Ducreux, L.J.; Morris, J.A.; Usman, M.; Verrall, S.R.; Fuller, J.; Simpson, C.G.; Zhang, R.; Hedley, P.E.; et al. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant Cell Environ. 2014, 37, 439–450. [Google Scholar] [CrossRef]
- Wolf, S.; Marani, A.; Rudich, J. Effect of temperature on carbohydrate metabolism in potato plants. J. Exp. Bot. 1991, 42, 619–625. [Google Scholar] [CrossRef]
- Molteberg, E.L. Influence of soil characteristics on skin finish of Norwegian potatoes. In Proceedings of the 20th Triennial EAPR Conference, Versaille, France, 9–14 July 2017. [Google Scholar]
- George, T.S.; Taylor, M.A.; Dodd, I.C.; White, P.J. Climate Change and Consequences for Potato Production: A Review of Tolerance to Emerging Abiotic Stress. Potato Res. 2018, 60, 239–268. [Google Scholar] [CrossRef]
- Rensink, W.A.; Iobst, S.; Hart, A.; Stegalkina, S.; Buell, C.R. Gene expression profiling of potato responses to cold, heat, and salt stress. Funct. Integr. Genom. 2005, 5, 201–207. [Google Scholar] [CrossRef]
- Gangadhar, B.H.; Yu, J.W.; Sajeesh, K.; Park, S.W. A systematic exploration of high-temperature stress-responsive genes in potato using large-scale yeast functional screening. Mol. Genet. Genom. 2014, 289, 185–201. [Google Scholar] [CrossRef]
- Ginzberg, I.; Barel, G.; Ophir, R.; Tzin, E.; Tanami, Z.; Muddarangappa, T.; de Jong, W.; Fogelman, E. Transcriptomic profiling of heat-stress response in potato periderm. J. Exp. Bot. 2009, 60, 4411–4421. [Google Scholar] [CrossRef]
- Hastilestari, B.R.; Lorenz, J.; Reid, S.; Hofmann, J.; Pscheidt, D.; Sonnewald, U.; Sonnewald, S. Deciphering source and sink responses of potato plants (Solanum tuberosum L.) to elevated temperatures. Plant Cell Environ. 2018, 41, 2600–2616. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fu, Q.; Huang, W.; Yu, D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009, 28, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, J.; Srivastava, R.; Bassham, D.C.; Howell, S.H. The Transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 2020, 32, 3559–3575. [Google Scholar] [CrossRef]
- Li, W.; Dong, J.; Cao, M.; Gao, X.; Wang, D.; Liu, B.; Chen, Q. Genome-wide identification and characterization of HD-ZIP genes in potato. Gene 2019, 697, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef]
- Sprenger, H.; Kurowsky, C.; Horn, R.; Erban, A.; Seddig, S.; Rudack, K.; Fischer, A.; Walther, D.; Zuther, E.; Köhl, K.; et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016, 39, 2370–2389. [Google Scholar] [CrossRef]
- Veselova, S.V.; Farkhutdinov, R.G.; Veselov, D.S.; Kudoyarova, G.R. Role of cytokinins in the regulation of stomatal conductance of wheat seedlings under conditions of rapidly changing local temperature. Russ. J. Plant Physiol. 2006, 53, 756–761. [Google Scholar] [CrossRef]
- Baker, N.R. Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [Green Version]
- Lahr, E.C.; Schade, G.W.; Crossett, C.C.; Watson, M.R. Photosynthesis and isoprene emission from trees along an urban-rural gradient in Texas. Glob. Chang. Biol. 2015, 21, 4221–4236. [Google Scholar] [CrossRef]
- Lafta, A.H.; Lorenzen, J.H. Effect of high temperature on plant growth and carbohydrate metabolism in potato. Plant Physiol. 1995, 109, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Pang, X.; Cheng, Y.; Yin, Y.; Zhang, Q.; Su, W.; Hu, B.; Guo, Q.; Ha, S.; Zhang, J.; et al. The Hsp70 Gene Family in Solanum tuberosum: Genome-Wide Identification, Phylogeny, and Expression Patterns. Sci. Rep.-UK 2018, 8, 16628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.R.; Lee, G.J.; Vierling, E. Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 1996, 47, 325–338. [Google Scholar] [CrossRef]
- Ahn, Y.J.; Claussen, K.; Zimmerman, J.L. Genotypic differences in the heat-shock response and thermotolerance in four potato cultivars. Plant Sci. 2004, 166, 901–911. [Google Scholar] [CrossRef]
- Ahn, Y.J.; Zimmerman, J.L. Introduction of the carrot HSP17.7 into potato (Solanum tuberosum L.) enhances cellular membrane stability and tuberization in vitro. Plant Cell Environ. 2006, 29, 95–104. [Google Scholar] [CrossRef]
- Trapero-Mozos, A.; Morris, W.L.; Ducreux, L.J.M.; McLean, K.; Stephens, J.; Torrance, L.; Bryan, G.J.; Hancock, R.D.; Taylor, M.A. Engineering heat tolerance in potato by temperature-dependent expression of a specific allele of HEAT-SHOCK COGNATE 70. Plant Biotech. J. 2018, 16, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Craig, E.A.; Huang, P.; Aron, R.; Andrew, A. The diverse roles of J proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Bioch. P 2006, 156, 1–21. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Savić, J.; Dragićević, I.; Pantelić, D.; Oljača, J.; Momčilović, I. Expression of small heat shock proteins and heat tolerance in potato (Solanum tuberosum L.). Arch. Biol. Sci. 2012, 64, 135–144. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Zu, Y.G.; Tang, Z.H.; Yu, J.H.; Liu, S.G.; Wang, W.; Guo, X.R. Different responses of camptothecin and 10-hydroxycamptothecin to heat shock in Camptotheca acuminate seedlings. Acta Bot. Sin. 2003, 45, 809–814. [Google Scholar]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Ruiz, J.M.; García, P.C.; López-Lefebre, L.R.; Sánchez, E.; Romero, L. Resistance to cold and heat stress: Accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 2001, 160, 315–321. [Google Scholar] [CrossRef]
- Martinez, V.; Mestre, T.C.; Rubio, F.; Girones-Vilaplana, A.; Moreno, D.A.; Mittler, R.; Rivero, R.M. Accumulation of flavonols over hydroxycinnamic acids favors oxidative damage protection under abiotic stress. Front. Plant Sci. 2016, 7, 838. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Ghazanfar, A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. J. Plant Physiol. 2006, 163, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Watkinson, J.I.; Juan, M.R.; Pablo, C.G.; Luis, R.L.; Esteban, S.; Luis, R. Accessions of Solanum tuberosum ssp. andigena show differences in photosynthetic recovery after drought stress as reflected in gene expression profiles. Plant Sci. 2006, 171, 745–758. [Google Scholar] [CrossRef]
- Simon-Sarkadi, L.; Kocsy, G.; Várhegyi, Á.; Galiba, G.; De Ronde, J.A. Genetic manipulation of proline accumulation influences the concentrations of other amino acids in soybean subjected to simultaneous drought and heat stress. J. Agric. Food Chem. 2005, 53, 7512–7517. [Google Scholar] [CrossRef]
- Chu, T.M.; Aspinall, D.; Paleg, L.G. Stress metabolism. VI. Temperature stress and the accumulation of proline in barley and radish. Aust. J. Plant Physiol. 1974, 1, 87–97. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.J.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense pathways collide: The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Lv, W.T.; Lin, B.; Zhang, M.; Hua, X.J. Proline accumulation is inhibitory to Arabidopsis seedlings during heat stress. Plant Physiol. 2011, 156, 1921–1933. [Google Scholar] [CrossRef] [Green Version]
- Evers, D.; Lefèvre, I.; Legay, S.; Lamoureux, D.; Hausman, J.F.; Rosales, R.O.; Marca, L.R.; Hoffmann, L.; Bonierbale, M.; Schafleitner, R. Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J. Exp Bot. 2010, 61, 2327–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Inskeep, W.P.; Bloom, P.R. Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physol. 1985, 77, 483–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zhang, G.; Murphy, A.; De Koeyer, D.; Tai, H.; Bizimungu, B.; Si, H.; Li, X.Q. Differences between the bud end and stem end of potatoes in dry matter content, starch granule size, and carbohydrate metabolicgene expression at the growing and sprouting stages. J. Agric. Food Chem. 2016, 64, 1176–1184. [Google Scholar] [CrossRef]



| H6h | Overlap | H3d | |
|---|---|---|---|
| Transcriptome | |||
| DEG up | 160 | 157 | 130 |
| DEG down | 538 | 285 | 94 |
| TF up | 14 | 6 | 7 |
| TF down | 56 | 30 | 14 |
| Metabolome (−)LC-MS | |||
| Compound up | 24 | 12 | 86 |
| Compound down | 15 | 10 | 94 |
| Metabolome (+)LC-MS | |||
| Compound up | 10 | 3 | 115 |
| Compound down | 44 | 32 | 156 |
| ID | Formula | Kegg_ID | Pathway Name | log2FC | p Value | VIP |
|---|---|---|---|---|---|---|
| H6h vs. HC | ||||||
| Com_99_neg | C38H60O18 | cpd:C09189 | Biosynthesis of secondary metabolites | −1.56891 | 0.032739 | 1.946182 |
| Com_224_neg | C26H28O16 | cpd:C12637 | Flavone and flavonol biosynthesis | 1.188334 | 0.013747 | 1.470693 |
| Com_232_neg | C8H9NO3 | cpd:C00250 | Metabolic pathways | 1.297375 | 0.032787 | 1.604524 |
| Com_608_neg | C5H9NO2 | cpd:C00148 | Biosynthesis of amino acids | 2.598589 | 0.00482 | 3.211703 |
| H3d vs. HC | ||||||
| Com_330_neg | C16H30O2 | cpd:C08362 | Fatty acid biosynthesis | −3.80026 | 0.000541 | 2.634698 |
| Com_1053_neg | C27H44O3 | cpd:C01673 | Steroid biosynthesis | −3.62372 | 0.002722 | 2.513606 |
| Com_22_neg | C8H10O2 | cpd:C06044 | Tyrosine metabolism | 2.141449 | 0.024333 | 1.481691 |
| Com_605_neg | C28H44N2O8S | cpd:C06462 | Arachidonic acid metabolism | 1.843223 | 0.005207 | 1.278471 |
| Com_224_neg | C26H28O16 | cpd:C12637 | Flavone and flavonol biosynthesis | 1.770409 | 0.025374 | 1.228481 |
| Com_1186_neg | C26H28O14 | cpd:C04858 | Flavone and flavonol biosynthesis | 1.644199 | 0.031581 | 1.137737 |
| Com_1450_pos | C3H7O6P | cpd:C00118 | Glycolysis/Gluconeogenesis | −1.7076 | 0.033928 | 1.322058 |
| Com_1193_pos | C6H9N3O2 | cpd:C00135 | Biosynthesis of amino acids | −1.48884 | 0.021547 | 1.149797 |
| Com_566_pos | C2H5O4P | cpd:C03167 | Phosphonate and phosphinate metabolism | −2.13038 | 0.002935 | 1.646439 |
| Com_2095_pos | C30H54N10O10S2 | cpd:C16564 | Glutathione metabolism | −1.92443 | 0.010559 | 1.488118 |
| Com_3261_pos | C18H32O4 | cpd:C04717 | Linoleic acid metabolism | −3.14728 | 0.000754 | 2.432073 |
| Com_1277_pos | C12H18O3 | cpd:C08491 | Plant hormone signal transduction | −1.32988 | 0.003432 | 1.028105 |
| Com_1814_pos | C24H29NO10 | cpd:C11813 | Isoquinoline alkaloid biosynthesis | −1.42629 | 0.009259 | 1.101574 |
| Com_1804_pos | C21H30O3 | cpd:C03205 | Metabolic pathways | 1.348268 | 0.046435 | 1.038995 |
| Com_150_pos | C7H8 | cpd:C01455 | Metabolic pathways | 3.252694 | 0.037825 | 2.494752 |
| Com_517_pos | C21H30O2 | cpd:C00410 | Metabolic pathways | 1.928316 | 0.035943 | 1.501117 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Kong, L.; Zhang, Y.; Liao, Y. Gene and Metabolite Integration Analysis through Transcriptome and Metabolome Brings New Insight into Heat Stress Tolerance in Potato (Solanum tuberosum L.). Plants 2021, 10, 103. https://doi.org/10.3390/plants10010103
Liu B, Kong L, Zhang Y, Liao Y. Gene and Metabolite Integration Analysis through Transcriptome and Metabolome Brings New Insight into Heat Stress Tolerance in Potato (Solanum tuberosum L.). Plants. 2021; 10(1):103. https://doi.org/10.3390/plants10010103
Chicago/Turabian StyleLiu, Bailin, Lingshuang Kong, Yu Zhang, and Yuncheng Liao. 2021. "Gene and Metabolite Integration Analysis through Transcriptome and Metabolome Brings New Insight into Heat Stress Tolerance in Potato (Solanum tuberosum L.)" Plants 10, no. 1: 103. https://doi.org/10.3390/plants10010103
APA StyleLiu, B., Kong, L., Zhang, Y., & Liao, Y. (2021). Gene and Metabolite Integration Analysis through Transcriptome and Metabolome Brings New Insight into Heat Stress Tolerance in Potato (Solanum tuberosum L.). Plants, 10(1), 103. https://doi.org/10.3390/plants10010103




