Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm
Abstract
:1. Introduction
2. Plant Holobiont
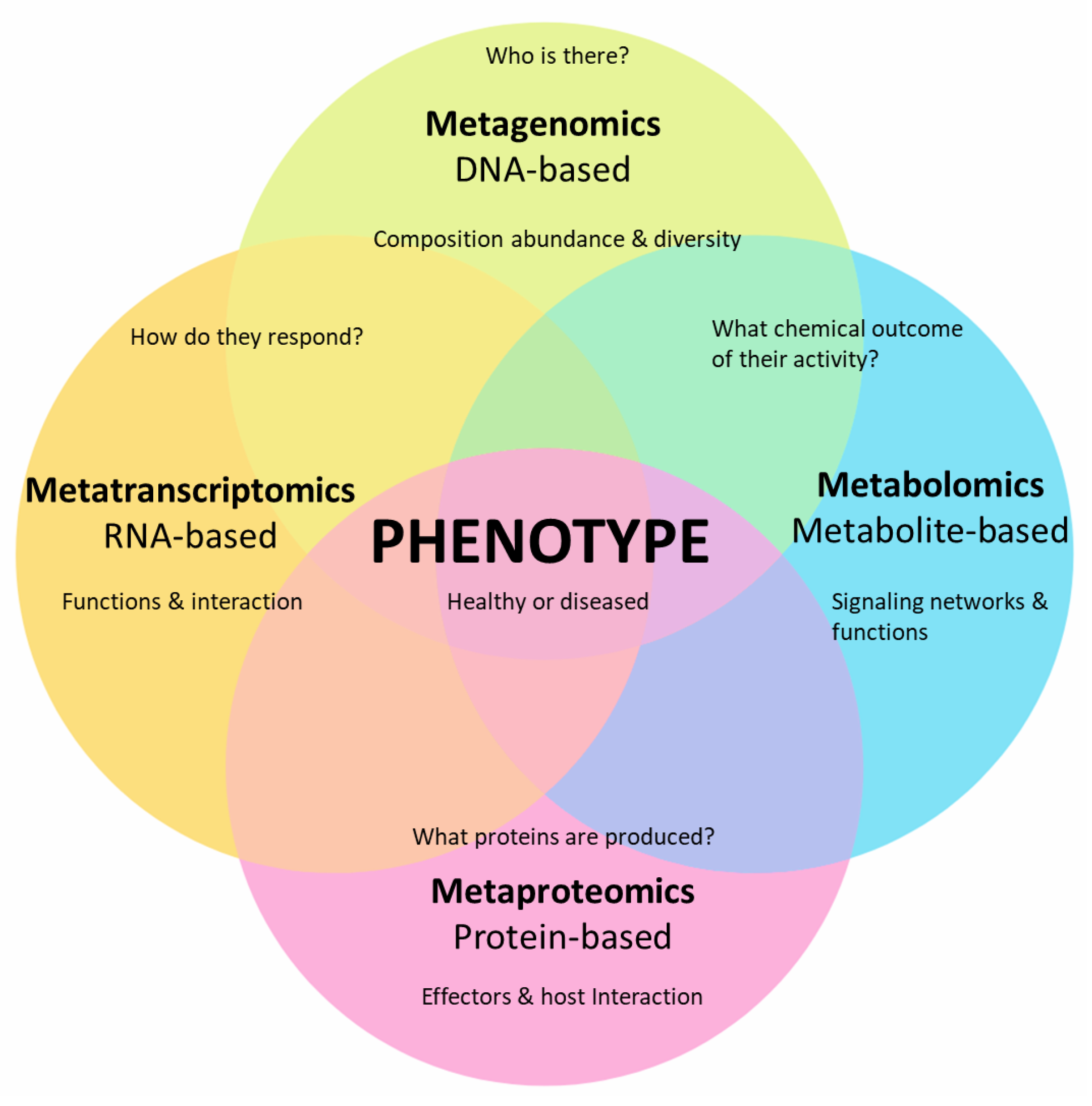
3. Meta-Omics and Plant Phytobiome Studies
4. Metabolomics and Modern Plant Pathology
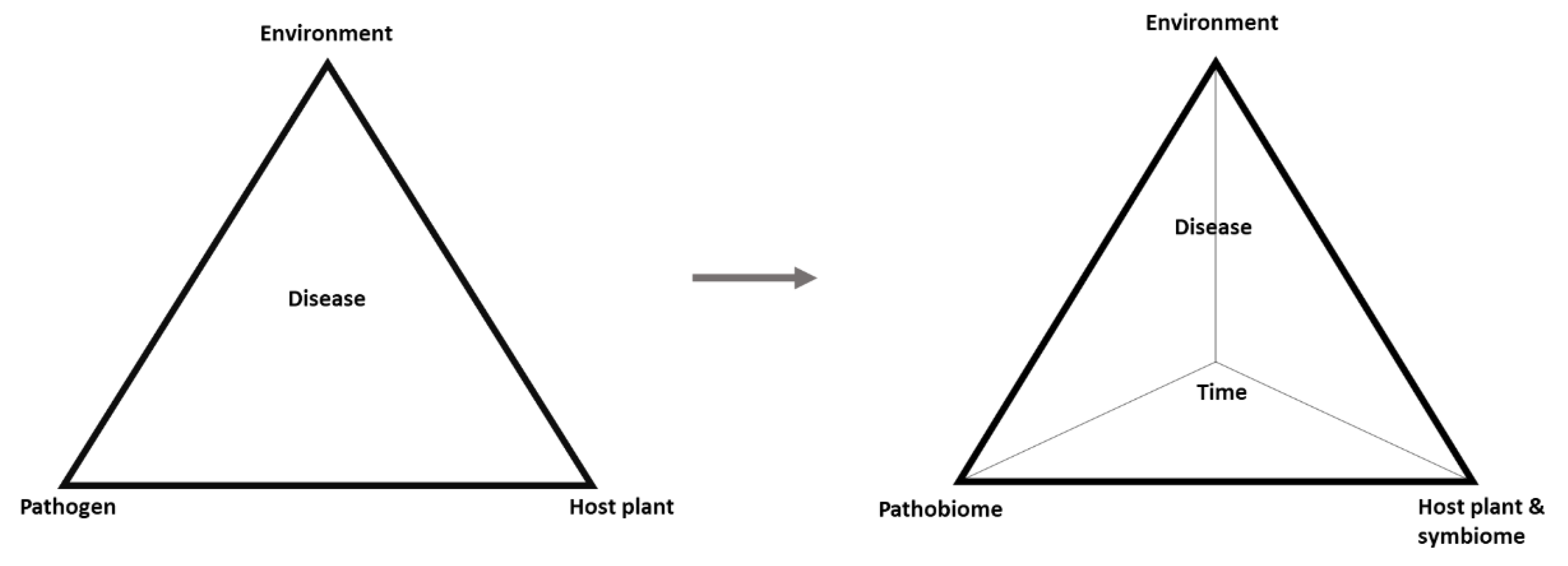
5. Plant Pathobiome
5.1. Co-Infections by and Interactions amongst Pathogenic Agents
5.2. Development and Assembly of the Pathobiome
6. Case Studies of Model Diseases Studied from the Pathobiome Perspective
6.1. Acute Oak Decline from the Perspective of the Pathobiome
6.2. Pine Wilt Disease from the Perspective of the Pathobiome
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bettenfeld, P.; Fontaine, F.; Trouvelot, S.; Fernandez, O.; Courty, P.E. Woody Plant Declines. What’s wrong with the Microbiome? Trends Plant Sci. 2020, 25, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Vayssier-Taussat, M.; Albina, E.; Citti, C.; Cosson, J.; Jacques, M.-A.; Lebrun, M.-H.; Le Loir, Y.; Ogliastro, M.; Petit, M.-A.; Roumagnac, P.; et al. Shifting the paradigm from pathogens to pathobiome: New concepts in the light of meta-omics. Front. Cell Infect. Microbiol. 2014, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltrus, D.A. Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol. 2017, 38, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Little, A.E.; Robinson, C.J.; Peterson, S.B.; Raffa, K.F.; Handelsman, J. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 2008, 62, 375–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, J.; Youle, M.; Knowlton, N.; Rohwer, F.; Relman, D.A. Superorganisms and Holobionts. Microbe 2013, 8, 152–153. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remusemsermann, M.N.P.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nat. Cell Biol. 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; DeAngelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G. The disease triangle: Pathogens, the environment and society. Nat. Rev. Genet. 2006, 5, 152–156. [Google Scholar] [CrossRef]
- Ogoshi, A. Introduction—The genus Rhizoctonia. In Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Sneh, B., Jabaji-hare, S., Neate, S., Dijst, G., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 1–9. [Google Scholar]
- Wu, J.; Ma, H.; Lü, M.; Han, S.; Zhu, Y.; Jin, H.; Liang, J.; Liu, L.; Xu, J. Rhizoctonia fungi enhance the growth of the endangered orchid Cymbidium goeringii. Botany 2010, 88, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Buonaurio, R.; Moretti, C.; Da Silva, D.P.; Cortese, C.; Ramos, C.; Venturi, V. The olive knot disease as a model to study the role of interspecies bacterial communities in plant disease. Front. Plant Sci. 2015, 6, 434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosni, T.; Moretti, C.; Devescovi, G.; Suarez-Moreno, Z.R.; Fatmi, M.B.; Guarnaccia, C.; Pongor, S.; Onofri, A.; Buonaurio, R.; Venturi, V. Sharing of quorum-sensing signals and role of interspecies communities in a bacterial plant disease. ISME J. 2011, 5, 1857–1870. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, D.P.; Castañeda-Ojeda, M.P.; Moretti, C.; Buonaurio, R.; Ramos, C.; Venturi, V. Bacterial multispecies studies and microbiome analysis of a plant disease. Microbiology 2014, 160, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Marchi, G.; Sisto, A.; Cimmino, A.; Andolfi, A.; Cipriani, M.G.; Evidente, A.; Surico, G. Interaction between Pseudomonas savastanoi pv. savastanoi and Pantoea agglomerans in olive knots. Plant Pathol. 2006, 55, 614–624. [Google Scholar] [CrossRef]
- Kim, N.; Kim, J.J.; Kim, I.; Mannaa, M.; Park, J.; Kim, J.; Lee, H.H.; Lee, S.B.; Park, D.S.; Sul, W.J.; et al. Type VI secretion systems of plantpathogenic Burkholderia glumae BGR1 play a functionally distinct role in interspecies interactions and virulence. Mol. Plant Pathol. 2020, 21, 1055–1069. [Google Scholar] [CrossRef]
- Tian, B.-Y.; Cao, Y.; Zhang, K.-Q. Metagenomic insights into communities, functions of endophytes and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 2015, 5, 17087. [Google Scholar] [CrossRef] [Green Version]
- Cocolin, L.; Mataragas, M.; Bourdichon, F.; Doulgeraki, A.; Pilet, M.F.; Jagadeesan, B.; Rantsiou, K.; Phister, T. Next generation microbiological risk assessment metaomics: The next need for integration. Int. J. Food Microbiol. 2018, 287, 10–17. [Google Scholar] [CrossRef]
- Pervaiz, T.; Lotfi, A.; Haider, M.S.; Haifang, J.; Fang, J. High Throughput Sequencing Advances and Future Challenges. J. Plant Biochem. Physiol. 2017, 5, 1–8. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Abrantes, I.; Caracciolo, A.B.; Bevivino, A.; Ciancio, A.; Grenni, P.; Hrynkiewicz, K.; Kredics, L.; Proença, D.N. Belowground Microbiota and the Health of Tree Crops. Front. Microbiol. 2018, 9, 1006. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broberg, M.; Doonan, J.; Mundt, F.; Denman, S.; McDonald, J.E. Integrated multi-omic analysis of host-microbiota interactions in acute oak decline. Microbiome 2018, 6, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, T.; Zhang, K.; Zhu, X.; Guan, L.; Jiu, S.; Li, X.; Nasim, M.; Jia, H.; Fang, J. Integrated metatranscriptome and transcriptome reveals the microbial community composition and physiological function of xylem sap on grapevine during bleeding period. Genes Genom. 2019, 41, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Prabha, R.; Gupta, V.K.; Verma, M.K. Metatranscriptome Analysis Deciphers Multifunctional Genes and Enzymes Linked With the Degradation of Aromatic Compounds and Pesticides in the Wheat Rhizosphere. Front. Microbiol. 2018, 9, 1331. [Google Scholar] [CrossRef]
- Xu, T.; Lei, L.; Shi, J.; Wang, X.; Chen, J.; Xue, M.; Sun, S.; Zhan, B.; Xia, Z.; Jiang, N.; et al. Characterization of maize translational responses to sugarcane mosaic virus infection. Virus Res. 2019, 259, 97–107. [Google Scholar] [CrossRef]
- Asselin, J.E.; Lin, J.; Perez-Quintero, A.L.; Gentzel, I.; Majerczak, D.; Opiyo, S.O.; Zhao, W.; Paek, S.M.; Kim, M.G.; Coplin, D.L.; et al. Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii. Plant Physiol. 2015, 167, 1117–1135. [Google Scholar] [CrossRef] [Green Version]
- Castro-Moretti, F.R.; Gentzel, I.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant-Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Sonnewald, U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017, 90, 839–855. [Google Scholar] [CrossRef] [Green Version]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Zhou, Y.; Li, X.; Zhao, J.; Guo, N.; Xing, H. Metabolomics Analysis of Soybean Hypocotyls in Response to Phytophthora sojae Infection. Front. Plant Sci. 2018, 9, 1530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apparecido, R.D.P.; Carlos, E.F.; Lião, L.; Vieira, L.G.E.; Alcantara, G.B. NMR-based metabolomics of transgenic and non-transgenic sweet orange reveals different responses in primary metabolism during citrus canker development. Metabolomics 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef]
- Balendres, M.A.; Nichols, D.S.; Tegg, R.S.; Wilson, C.R. Metabolomes of Potato Root Exudates: Compounds That Stimulate Resting Spore Germination of the Soil-Borne Pathogen Spongospora subterranea. J. Agric. Food Chem. 2016, 64, 7466–7474. [Google Scholar] [CrossRef]
- Malinowski, R.; Nisler, J.; Borhan, M.H.; Spíchal, L.; Strnad, M.; Rolfe, S.A. The role of cytokinins in clubroot disease. Eur. J. Plant Pathol. 2016, 145, 543–557. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-K.; Ahn, S.; Cho, H.Y.; Yun, H.Y.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci. Rep. 2016, 6, 37434. [Google Scholar] [CrossRef] [Green Version]
- Collemare, J.; O’Connell, R.J.; Lebrun, M.-H. Nonproteinaceous effectors: The terra incognita of plant–fungal interactions. New Phytol. 2019, 223, 590–596. [Google Scholar] [CrossRef] [Green Version]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Genet. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Dallery, J.F.; Lapalu, N.; Zampounis, A.; Pigné, S.; Luyten, I.; Amselem, J.; Wittenberg, A.H.J.; Zhou, S.; de Queiroz, M.V.; Robin, G.P.; et al. Gapless genome assembly of Colletotrichum higginsianum reveals chromosome structure and association of tranposable elements with secondary metabolite gene clusters. BMC Genom. 2017, 18, 667. [Google Scholar] [CrossRef]
- Harris, L.J.; Balcerzak, M.; Johnston, A.; Schneiderman, D.; Ouellet, T. Host preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016, 120, 111–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiberg, A.; Wang, M.; Lin, F.-M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.-D.; Jin, H. Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesisrelated 2 gene. New Phytol. 2017, 215, 338–350. [Google Scholar] [CrossRef] [Green Version]
- De Fazio, J.; Fleming, I.D.; Shakhsheer, B.; Zaborina, O.; Alverdy, J.C. The opposing forces of the intestinal Microbiome and the emerging pathobiome. Surg. Clin. N. Am. 2014, 94, 1151–1161. [Google Scholar] [CrossRef] [Green Version]
- Bass, D.; Stentiford, G.D.; Wang, H.-C.; Koskella, B.; Tyler, C.R. The Pathobiome in Animal and Plant Diseases. Trends Ecol. Evol. 2019, 34, 996–1008. [Google Scholar] [CrossRef] [Green Version]
- Pitlik, S.D.; Koren, O. How holobionts get sick—Toward a unifying scheme of disease. Microbiome 2017, 5, 1–4. [Google Scholar] [CrossRef]
- Whitelaw-Weckert, M.A.; Rahman, L.; Appleby, L.M.; Hall, A.; Clark, A.C.; Waite, H.; Hardie, W.J. Coinfection by Botryosphaeriaceae and Ilyonectria spp. fungi during propagation causes decline of young grafted grapevines. Plant Pathol. 2013, 62, 1226–1237. [Google Scholar] [CrossRef]
- Moura, M.L.; Jacques, L.A.; Brito, L.M.; Mourao, I.M.; Duclos, J. Tomato pith necrosis caused by P. corrugata and P. mediterranea: Severity of damages and crop loss assessment. Acta Hort. 2005, 695, 365–372. [Google Scholar] [CrossRef]
- Canaday, C.H.; Wyatt, J.E.; Mullins, J.A. Resistance to broccoli to bacterial soft rot caused by Pseudomonas marginalis and fluorescent Pseudomonas species. Plant Dis. 1991, 75, 715–720. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Venturi, V. Synergisms between microbial pathogens in plant disease complexes: A growing trend. Front. Plant Sci. 2015, 6, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, B.; Park, J.; Kim, N.; Li, T.; Kim, S.; Bartley, L.E.; Kim, J.; Kim, I.; Kang, Y.; Yun, K.; et al. Cooperative interactions between seed-borne bacterial and air-borne fungal pathogens on rice. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stopnisek, N.; Zühlke, D.; Carlier, A.; Barberán, A.; Fierer, N.; Becher, D.; Riedel, K.; Eberl, L.; Weisskopf, L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2015, 10, 253–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partida-Martinez, L.P.; Hertweck, C. A Gene Cluster Encoding Rhizoxin Biosynthesis in “Burkholderia rhizoxina”, the Bacterial Endosymbiont of the Fungus Rhizopus microsporus. ChemBioChem 2006, 8, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Iwasaki, S.; Kobayashi, H.; Okuda, S.; Murai, T.; Sato, Y. Rhizoxin binding to tubulin at the maytansine-binding site. Biochim. Biophys. Acta Gen. Subj. 1987, 926, 215–223. [Google Scholar] [CrossRef]
- Scherlach, K.; Busch, B.; Lackner, G.; Paszkowski, U.; Hertweck, C. Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. 2012, 124, 9753–9756. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nat. Cell Biol. 2005, 437, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Lackner, G.; Moebius, N.; Hertweck, C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2010, 5, 252–261. [Google Scholar] [CrossRef] [Green Version]
- Lackner, G.; Moebius, N.; Partida-Martínez, L.P.; Boland, S.; Hertweck, C. Evolution of an endofungal Lifestyle: Deductions from the Burkholderia rhizoxinica Genome. BMC Genom. 2011, 12, 210–213. [Google Scholar] [CrossRef] [Green Version]
- Cooper, A.J.; Latunde-Dada, A.O.; Woods-Tör, A.; Lynn, J.; Lucas, J.A.; Crute, I.R.; Holub, E.B. Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad spectrum suppression of innate immunity. Mol. Plant Microbe Interact. 2008, 21, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Prince, D.C.; Rallapalli, G.; Xu, D.; Schoonbeek, H.J.; Çevik, V.; Asai, S.; Kemen, E.; Cruz-Mireles, N.; Kemen, A.; Belhaj, K.; et al. Albugo-imposed changes to tryptophanderived antimicrobial metabolite biosynthesis may contribute to suppression of non-host resistance to Phytophthora infestans in Arabidopsis thaliana. BMC Biol. 2017, 15, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.; Mannaa, M.; Kim, J.; Lee, C.; Kim, S.M.; Ra, J.E.; Lee, H.H.; Seo, Y.S. The in vitro and in planta interspecies interactions among rice-pathogenic Burkholderia species. Plant Dis. 2020, PDIS-06. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, H. Biocontrol of bacterial seedling blight of rice caused by Burkholderia gladioli using with its avirulent isolate. Jpn. J. Phytopathol. 2000, 66, 232–238. [Google Scholar] [CrossRef]
- Riera-Ruiz, C.; Castro-Lara, J.; Jimenez-Feijoó, M.I.; Cevallos-Cevallos, J.M. Interactions of Burkholderia glumae and B. gladioli in symptom development in rice seeds and seedlings. Can. J. Plant Pathol. 2018, 40, 347–357. [Google Scholar] [CrossRef]
- Elshafie, H.S.; Racioppi, R.; Bufo, S.A.; Camele, I. In vitro study of biological activity of four strains of Burkholderia gladioli pv. agaricicola and identification of their bioactive metabolites using GC-MS. Saudi J. Biol. Sci. 2016, 24, 295–301. [Google Scholar] [CrossRef] [Green Version]
- De Wit, R.; Bouvier, T. Everything is everywhere, but, the environment selects, what did Baas Becking and Beijerinck really say? Environ. Microbiol. 2006, 8, 755–758. [Google Scholar] [CrossRef]
- Mannaa, M.; Park, I.; Seo, Y.-S. Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species. Int. J. Mol. Sci. 2018, 20, 121. [Google Scholar] [CrossRef] [Green Version]
- Partida-Martinez, L.P.; Monajembashi, S.; Greulich, K.O.; Hertweck, C. Endosymbiont-dependent host reproduction maintains bacterial fungal mutualism. Curr. Biol. 2007, 17, 773–777. [Google Scholar] [CrossRef] [Green Version]
- Hacker, J.; Kaper, J.B. Pathogenicity Islands and the Evolution of Microbes. Annu. Rev. Microbiol. 2000, 54, 641–679. [Google Scholar] [CrossRef] [Green Version]
- Juhas, M.; Van Der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef] [Green Version]
- Rogers, G.; Hoffman, L.R.; Carroll, M.P.; Bruce, K.D. Interpreting infective microbiota: The importance of an ecological perspective. Trends Microbiol. 2013, 21, 271–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denman, S.; Brown, N.; Kirk, S.; Jeger, M.; Webber, J. A description of the symptoms of Acute Oak Decline in Britain and a comparative review on causes of similar disorders on oak in Europe. For. Int. J. For. Res. 2014, 87, 535–551. [Google Scholar] [CrossRef] [Green Version]
- Ruffner, B.; Schneider, S.; Meyer, J.; Queloz, V.; Rigling, D. First report of acute oak decline disease of native and non-native oaks in Switzerland. New Dis. Rep. 2020, 41, 18. [Google Scholar] [CrossRef] [Green Version]
- Doonan, J.M.; Broberg, M.; Denman, S.; McDonald, J.E. Host microbiota insect interactions drive emergent virulence in a complex tree disease. Proc. R. Soc. B 2020, 287, 20200956. [Google Scholar] [CrossRef]
- Denman, S.; Doonan, J.; Ransom-Jones, E.; Broberg, M.; Plummer, S.; Kirk, S.; Scarlett, K.; Griffiths, A.R.; Kaczmarek, M.; Forster, J.; et al. Microbiome and infectivity studies reveal complex polyspecies tree disease in Acute Oak Decline. ISME J. 2018, 12, 386–399. [Google Scholar] [CrossRef] [Green Version]
- Pinho, D.; Barroso, C.; Froufe, H.; Brown, N.; Vanguelova, E.; Egas, C.; Denman, S. Linking tree health, rhizosphere physi-cochemical properties, and microbiome in acute oak decline. Forests 2020, 11, 1153. [Google Scholar] [CrossRef]
- Tóth, Á. Bursaphelenchus xylophilus, the pinewood nematode: Its significance and a historical review. Acta Biol. Szeged. 2011, 55, 213–217. [Google Scholar]
- Zhao, L.; Mota, M.; Vieira, P.; Butcher, R.A.; Sun, J. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014, 30, 299–308. [Google Scholar] [CrossRef] [Green Version]
- Futai, K. Pine wood nematode, Bursaphelenchus xylophilus. Ann. Rev. Phytopathol. 2013, 51, 61–83. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLOS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [Green Version]
- Shinya, R.; Morisaka, H.; Takeuchi, Y.; Futai, K.; Ueda, M. Making headway in understanding pine wilt disease: What do we perceive in the postgenomic era? J. Biosci. Bioeng. 2013, 116, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Shibuya, H.; Jones, J.T. Molecular and biochemical characterization of an endo-β-1,3-glucanase from the pinewood nematode Bursaphelenchus xylophilus acquired by horizontal gene transfer from bacteria. Biochem. J. 2005, 389, 117–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maehara, N.; Futai, K. Effect of fungal interactions on the numbers of the pinewood nematode, Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae), carried by the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae). Fundam. Appl. Nematol. 1997, 20, 611–618. [Google Scholar]
- Zhao, L.; Lu, M.; Niu, H.; Fang, G.; Zhang, S.; Sun, J. A native fungal symbiont facilitates the prevalence and development of an invasive pathogen-native vector symbiosis. Ecology 2013, 94, 2817–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamiya, Y. Scanning Electron Microscopy of Pine Seedling Wood Tissue Sections Inoculated with the Pinewood Nematode Bursaphelenchus xylophilus Previously Prepared for Light Microscopy. J. Nematol. 2012, 44, 255–259. [Google Scholar]
- Cheng, X.Y.; Tian, X.L.; Wang, Y.S.; Lin, R.M.; Mao, Z.C.; Chen, N.; Xie, B.Y. Metagenomic analysis of the pinewood nematode microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci. Rep. 2013, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Vicente, C.S.; Ikuyo, Y.; Mota, M.; Hasegawa, K. Pinewood nematode-associated bacteria contribute to oxidative stress resistance of Bursaphelenchus xylophilus. BMC Microbiol. 2013, 13, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Oku, H.; Shiraishi, T.; Ouchi, S.; Kurozumi, S.; Ohta, H. Pine wilt toxin, the metabolite of a bacterium associated with a nematode. Naturwissenschaften 1980, 67, 198–199. [Google Scholar] [CrossRef]
- Han, Z.; Hong, Y.D.; Zhao, B.G. A Study on Pathogenicity of Bacteria Carried by Pine Wood Nematodes. J. Phytopathol. 2003, 151, 683–689. [Google Scholar] [CrossRef]
- Le Dang, Q.; Son, S.W.; Cheon, H.-M.; Choi, G.J.; Choi, Y.H.; Jang, K.S.; Lim, C.H.; Kim, J.-C. Pyochelin isolated from Burkholderia arboris KRICT1 carried by pine wood nematodes exhibits phytotoxicity in pine callus. Nematology 2011, 13, 521–528. [Google Scholar] [CrossRef]
- Guo, Q.; Guo, D.; Zhao, B.; Xu, J.; Li, R. Two Cyclic Dipeptides from Pseudomonas fluorescens GcM5-1A Carried by the Pine Wood Nematode and Their Toxicities to Japanese Black Pine Suspension Cells and Seedlings in vitro. J. Nematol. 2007, 39, 243–247. [Google Scholar] [PubMed]
- Kawazu, K.; Zhang, H.; Yamashita, H.; Kanzaki, H. Relationship between the Pathogenicity of the Pine Wood Nematode, Bursaphelenchus xylophilus, and Phenylacetic Acid Production. Biosci. Biotechnol. Biochem. 1996, 60, 1413–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proença, D.N.; Grass, G.; Morais, P.V. Understanding pine wilt disease: Roles of the pine endophytic bacteria and of the bacteria carried by the disease-causing pinewood nematode. Microbiology 2016, 6, e00415. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.S.; Pereira, A.; Vicente, C.S.L.; Matos, P.; Henriques, J.; Lopes, H.; Nascimento, F.X.; Mota, M.; Correia, A.; Henriques, I. The role of bacteria in pine wilt disease: Insights from microbiome analysis. FEMS Microbiol. Ecol. 2018, 94, 077. [Google Scholar] [CrossRef] [PubMed]
- Mannaa, M.; Han, G.; Jeon, H.W.; Kim, J.; Kim, N.; Park, A.R.; Kim, J.-C.; Seo, Y.-S. Influence of Resistance-Inducing Chemical Elicitors against Pine Wilt Disease on the Rhizosphere Microbiome. Microorganisms 2020, 8, 884. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Jeon, H.W.; Mannaa, M.; Jeong, S.I.; Kim, J.; Kim, J.; Lee, C.; Park, A.R.; Kim, J.C.; Seo, Y.S. Induction of resistance against pine wilt disease caused by Bursaphelenchus xylophilus using selected pine endophytic bacteria. Plant Pathol. 2019, 68, 434–444. [Google Scholar] [CrossRef]

| Interacting Species | Host Plant and Disease | Type of Interaction |
|---|---|---|
| Burkholderia glumae ↔ Fusarium graminearum | Rice panicle and seedling blight—Fusarium head blight | Bacteria-fungi co-operative interaction |
| Mycetohabitans rhizoxinica ↔ Rhizopus microsporus | Rice seedling blight | Bacteria-fungi endosymbiotic mutualism |
| Albugo candida ↔ Phytophthora infestans | Crucifers, downy or powdery mildew and Phytophthora blight | Fungi-fungi co-operative interaction |
| Burkholderia gladioli ↔ Burkholderia glumae and Burkholderia plantarii | Rice panicle and seedling blight | Bacteria-bacteria antagonism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannaa, M.; Seo, Y.-S. Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm. Plants 2021, 10, 125. https://doi.org/10.3390/plants10010125
Mannaa M, Seo Y-S. Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm. Plants. 2021; 10(1):125. https://doi.org/10.3390/plants10010125
Chicago/Turabian StyleMannaa, Mohamed, and Young-Su Seo. 2021. "Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm" Plants 10, no. 1: 125. https://doi.org/10.3390/plants10010125
APA StyleMannaa, M., & Seo, Y.-S. (2021). Plants under the Attack of Allies: Moving towards the Plant Pathobiome Paradigm. Plants, 10(1), 125. https://doi.org/10.3390/plants10010125






