Genome-Wide Analysis of the COBRA-Like Gene Family Supports Gene Expansion through Whole-Genome Duplication in Soybean (Glycine max)
Abstract
:1. Introduction
2. Results
2.1. Identification of COBL Genes in Soybean
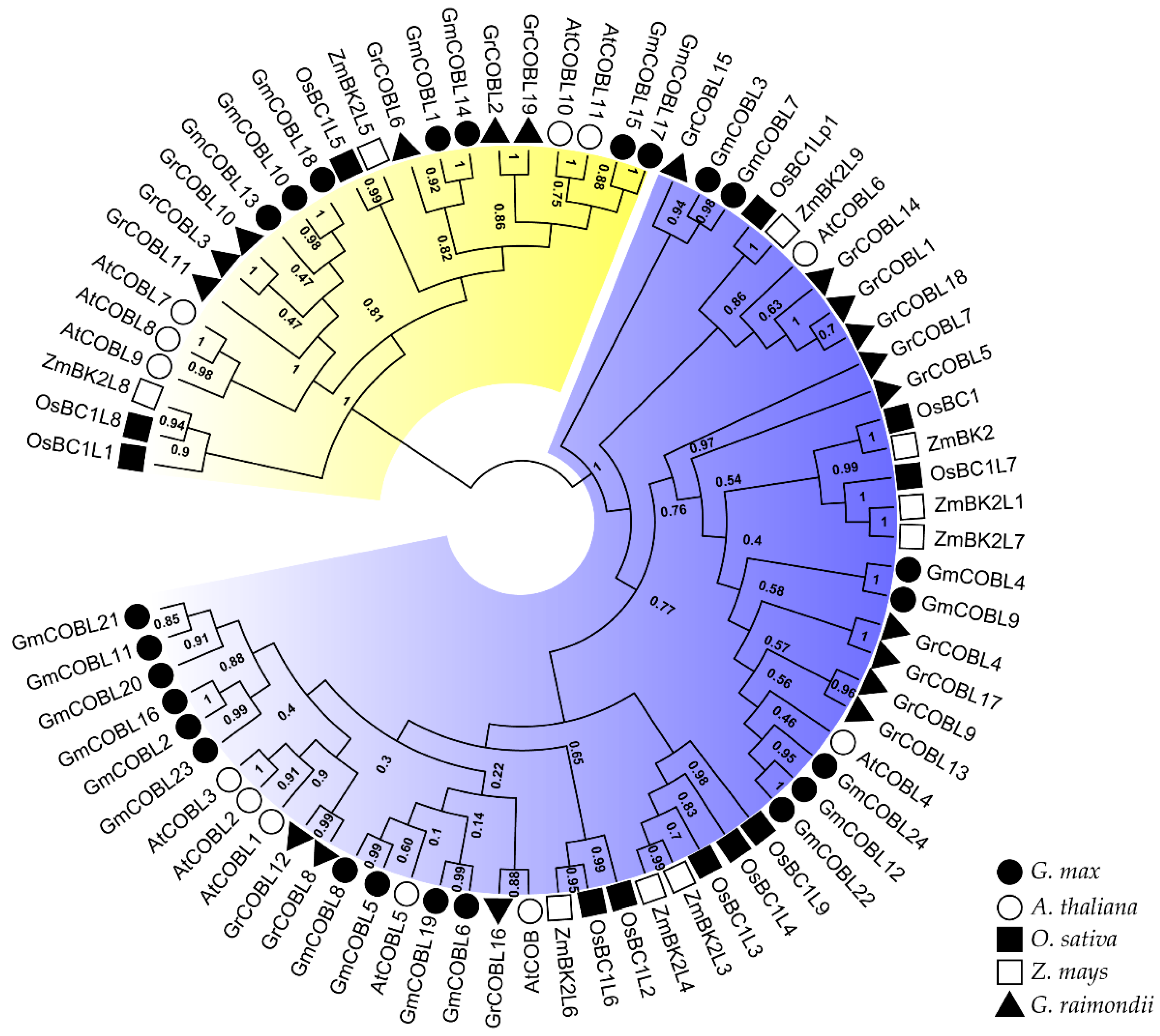
2.2. Phylogenetic and Structural Analysis of COBL Genes in Soybean
2.3. Distribution and Duplication of COBL Genes in the Soybean Genome
2.4. Tissue Expression Profiling and Biological Process
2.5. Promoter Cis-Regulatory Element Analysis
3. Discussion
3.1. Soybean Contains 24 COBL Genes
3.2. WGD Collaborated to COBL Gene Expansion in Soybean
3.3. Expression Profiles of COBL Gene Family in Soybean Showed Functional Diversity
4. Materials and Methods
4.1. Identification of COBL Gene Family Members in the Soybean Genome
4.2. Phylogenetic Analysis
4.3. Gene Structure and Protein Conserved Domains and Motifs
4.4. Chromosome Location and Gene Duplication
4.5. Estimation of Non-Synonymous and Synonymous Substitution Rates and Evaluation Divergence Time
4.6. Expression Profile Analysis of Soybean COBL Genes
4.7. Putative Promoter Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering land plants—Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.M. Cellulose Structure and Biosynthesis: What is in Store for the 21st Century? J. Polym. Sci. Part A Polym. Chem. 2004, 42, 487–495. [Google Scholar] [CrossRef]
- Malcolm Brown, R.; Saxena, I.M.; Kudlicka, K. Cellulose biosynthesis in higher plants. Trends Plant Sci. 1996, 1, 149–156. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Li, L.; Sun, Y.-H.; Chiang, V.L. The Cellulose Synthase Gene Superfamily and Biochemical Functions of Xylem-Specific Cellulose Synthase-Like Genes in Populus trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef] [Green Version]
- Persson, S.; Wei, H.; Milne, J.; Page, G.P.; Somerville, C.R. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. USA 2005, 102, 8633–8638. [Google Scholar] [CrossRef] [Green Version]
- Schindelman, G.; Morikami, A.; Jung, J.; Baskin, T.I.; Carpita, N.C.; Derbyshire, P.; McCann, M.C.; Benfey, P.N. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in arabidopsis. Genes Dev. 2001, 15, 1115–1127. [Google Scholar] [CrossRef] [Green Version]
- Roudier, F. The COBRA Family of Putative GPI-Anchored Proteins in Arabidopsis. A New Fellowship in Expansion. Plant Physiol. 2002, 130, 538–548. [Google Scholar] [CrossRef] [Green Version]
- Borner, G.H.H.; Sherrier, D.J.; Stevens, T.J.; Arkin, I.T.; Dupree, P. Prediction of Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis. A Genomic Analysis. Plant Physiol. 2002, 129, 486–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Shang-Guan, K.; Zhang, B.; Liu, X.; Yan, M.; Zhang, L.; Shi, Y.; Zhang, M.; Qian, Q.; Li, J.; et al. Brittle Culm1, a COBRA-Like Protein, Functions in Cellulose Assembly through Binding Cellulose Microfibrils. PLoS Genet. 2013, 9, e1003704. [Google Scholar] [CrossRef] [PubMed]
- Bahari, M.N.A.; Sakeh, N.M.; Abdullah, S.N.A.; Ramli, R.R.; Kadkhodaei, S. Transciptome profiling at early infection of Elaeis guineensis by Ganoderma boninense provides novel insights on fungal transition from biotrophic to necrotrophic phase. BMC Plant Biol. 2018, 18, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Niu, E.; Fang, S.; Shang, X.; Guo, W. Ectopic expression of GhCOBL9A, a cotton glycosyl-phosphatidyl inositol-anchored protein encoding gene, promotes cell elongation, thickening and increased plant biomass in transgenic Arabidopsis. Mol. Genet. Genom. 2018, 293, 1191–1204. [Google Scholar] [CrossRef]
- Cao, Y.; Tang, X.; Giovannoni, J.; Xiao, F.; Liu, Y. Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. BMC Plant Biol. 2012, 12, 211. [Google Scholar] [CrossRef] [Green Version]
- Brady, S.M.; Song, S.; Dhugga, K.S.; Rafalski, J.A.; Benfey, P.N. Combining Expression and Comparative Evolutionary Analysis. The COBRA Gene Family. Plant Physiol. 2007, 143, 172–187. [Google Scholar] [CrossRef] [Green Version]
- Ching, A.; Dhugga, K.S.; Appenzeller, L.; Meeley, R.; Bourett, T.M.; Howard, R.J. Brittle stalk 2 encodes a putative glycosylphosphatidylinositol- anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 2006, 224, 1174–1184. [Google Scholar] [CrossRef]
- Niu, E.; Shang, X.; Cheng, C.; Bao, J.; Zeng, Y.; Cai, C.; Du, X.; Guo, W. Comprehensive analysis of the COBRA-like (COBL) gene family in Gossypium identifies two COBLs potentially associated with fiber quality. PLoS ONE 2015, 10, e0145725. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Kang, B.G.; Osburn, L.D.; Cheng, Z.M. The COBRA gene family in Populus and gene expression in vegetative organs and in response to hormones and environmental stresses. Plant Growth Regul. 2009, 58, 211–223. [Google Scholar] [CrossRef]
- Li, S.; Ge, F.; Xu, M.; Zhao, X.; Huang, G.; Zhou, L.; Wang, J.; Kombrink, A. Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J. 2013, 74, 486–497. [Google Scholar] [CrossRef]
- Sindhu, A.; Langewisch, T.; Olek, A.; Multani, D.S.; McCann, M.C.; Vermerris, W.; Carpita, N.C.; Johal, G. Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 2007, 145, 1444–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Suzuki, R.; Nishikubo, N.; Takenouchi, S.; Ito, S.; Nakano, Y.; Nakaba, S.; Sano, Y.; Funada, R.; Kajita, S.; et al. Isolation of a novel cell wall architecture mutant of rice with defective Arabidopsis COBL4 ortholog BC1 required for regulated deposition of secondary cell wall components. Plant Signal. Behav. 2010, 5, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, F.B.; Moharana, K.C.; Almeida-Silva, F.; Gazara, R.K.; Pedrosa-Silva, F.; Coelho, F.S.; Grativol, C.; Venancio, T.M. Systematic analysis of 1298 RNA-Seq samples and construction of a comprehensive soybean (Glycine max) expression atlas. Plant J. 2020, 103, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genetics. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Wen, T.J.; Zimmermann, R.; Chimot-Marolle, P.; Da Costa, E.; Silva, O.; Bruce, W.; Lamkey, K.R.; Wienand, U.; Schnable, P.S. The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J. 2008, 54, 888–898. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.; Waneck, G.L. Glycosyl-phosphatidylinositol-anchored membrane proteins. J. Am. Soc. Nephrol. 1992, 3, 895–906. [Google Scholar]
- Pagel, J.; Walling, J.G.; Young, N.D.; Shoemaker, R.C.; Jackson, S.A. Segmental duplications within the Glycine max genome revealed by fluorescence in situ hybridization of bacterial artificial chromosomes. Genome 2004, 47, 764–768. [Google Scholar] [CrossRef]
- Shoemaker, R.C.; Schlueter, J.; Doyle, J.J. Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 2006, 9, 104–109. [Google Scholar] [CrossRef]
- Taylor, N.G. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 239–252. [Google Scholar] [CrossRef]
- Li, P.; Liu, Y.; Tan, W.; Chen, J.; Zhu, M.; Lv, Y.; Liu, Y.; Yu, S.; Zhang, W.; Cai, H. Brittle Culm 1 Encodes a COBRA-Like Protein Involved in Secondary Cell Wall Cellulose Biosynthesis in Sorghum. Plant Cell Physiol. 2018, 60, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Ben-Tov, D.; Abraham, Y.; Stav, S.; Thompson, K.; Loraine, A.; Elbaum, R.; de Souza, A.; Pauly, M.; Kieber, J.J.; Harpaz-Saad, S. COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in arabidopsis seed coat mucilage secretory cells. Plant Physiol. 2015, 167, 711–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Tov, D.; Idan-Molakandov, A.; Hugger, A.; Ben-Shlush, I.; Günl, M.; Yang, B.; Usadel, B.; Harpaz-Saad, S. The role of COBRA-LIKE 2 function, as part of the complex network of interacting pathways regulating Arabidopsis seed mucilage polysaccharide matrix organization. Plant J. 2018, 94, 497–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francoz, E.; Ranocha, P.; Burlat, V.; Dunand, C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015, 20, 515–524. [Google Scholar] [CrossRef]
- Dai, X.; You, C.; Wang, L.; Chen, G.; Zhang, Q.; Wu, C. Molecular characterization, expression pattern, and function analysis of the OsBC1L family in rice. Plant Mol. Biol. 2009, 71, 469–481. [Google Scholar] [CrossRef]
- Sangi, S.; Santos, M.L.C.; Alexandrino, C.R.; Da Cunha, M.; Coelho, F.S.; Ribeiro, G.P.; Lenz, D.; Ballesteros, H.; Hemerly, A.S.; Venâncio, T.M.; et al. Cell wall dynamics and gene expression on soybean embryonic axes during germination. Planta 2019, 250, 1325–1337. [Google Scholar] [CrossRef]
- Bellieny-Rabelo, D.; de Oliveira, E.A.G.; Ribeiro, E. da S.; Costa, E.P.; Oliveira, A.E.A.; Venancio, T.M. Transcriptome analysis uncovers key regulatory and metabolic aspects of soybean embryonic axes during germination. Sci. Rep. 2016, 6, 36009. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 00, 2.3.1–2.3.22. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, D.; Dudek, S.; Ritchie, M.D.; Pendergrass, S.A. Visualizing genomic information across chromosomes with PhenoGram. BioData Min. 2013, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Coneryz, J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]


| Gene Name | Gene Locus | CDS Length (bp 1) | Protein Length (aa 2) | Exon Number | Transcript Number | CCVS Motif | N-terminal Signal Peptide | Potential ω- Site Position |
|---|---|---|---|---|---|---|---|---|
| GmCOBL1 | Glyma.01G240200 | 1800 | 599 | 4 | 1 | Yes | yes | N(573) |
| GmCOBL2 | Glyma.02G196100 | 1110 | 369 | 5 | 1 | Yes | no | none |
| GmCOBL3 | Glyma.04G006100 | 1344 | 447 | 6 | 1 | Yes | yes | N(418) |
| GmCOBL4 | Glyma.04G160000 | 1377 | 458 | 6 | 1 | Yes | yes | N(429) |
| GmCOBL5 | Glyma.04G160100 | 1371 | 456 | 6 | 1 | Yes | yes | P(425) |
| GmCOBL6 | Glyma.05G019100 | 630 | 209 | 5 | 1 | No | yes | none |
| GmCOBL7 | Glyma.06G005800 | 1338 | 445 | 6 | 1 | Yes | yes | G(420) |
| GmCOBL8 | Glyma.06G205400 | 1371 | 456 | 6 | 2 | Yes | yes | P(425) |
| GmCOBL9 | Glyma.06G205500 | 1377 | 458 | 6 | 1 | Yes | yes | N(429) |
| GmCOBL10 | Glyma.07G243200 | 1932 | 643 | 2 | 1 | Yes | yes | V(617) |
| GmCOBL11 | Glyma.08G249700 | 1347 | 448 | 6 | 1 | Yes | yes | N(422) |
| GmCOBL12 | Glyma.08G249800 | 1296 | 431 | 6 | 2 | Yes | yes | N(408) |
| GmCOBL13 | Glyma.09G039900 | 1953 | 650 | 2 | 1 | Yes | yes | S(625) |
| GmCOBL14 | Glyma.11G003300 | 1956 | 651 | 2 | 1 | Yes | yes | N(628) |
| GmCOBL15 | Glyma.12G213400 | 2016 | 671 | 2 | 1 | Yes | yes | S(647) |
| GmCOBL16 | Glyma.13G053300 | 1377 | 458 | 6 | 2 | Yes | yes | N(432) |
| GmCOBL17 | Glyma.13G288300 | 2001 | 666 | 2 | 1 | Yes | yes | S(642) |
| GmCOBL18 | Glyma.17G030700 | 1935 | 644 | 2 | 1 | Yes | yes | S(615) |
| GmCOBL19 | Glyma.17G080600 | 1365 | 454 | 6 | 1 | Yes | yes | A(423) |
| GmCOBL20 | Glyma.18G271900 | 1263 | 420 | 6 | 3 | No | yes | none |
| GmCOBL21 | Glyma.18G272000 | 1347 | 448 | 6 | 1 | Yes | yes | A(423) |
| GmCOBL22 | Glyma.18G272100 | 1296 | 431 | 6 | 2 | Yes | yes | N(408) |
| GmCOBL23 | Glyma.19G033500 | 1365 | 454 | 6 | 1 | Yes | yes | N(428) |
| GmCOBL24 | Glyma.19G033600 | 1296 | 431 | 6 | 2 | Yes | yes | N(408) |
| Species Name | Total Number of COBRA-Like Genes | Haploid Genome Size (Mb 1) | Chromosomes (n 2) | Reference |
|---|---|---|---|---|
| Glycine max | 24 | 1100 | 10 | Our study |
| Gossypium raimondii | 19 | 880 | 13 | (Niu, 2015) |
| Arabidopsis thaliana | 12 | 135 | 5 | (Roudier, 2002) |
| Oryza sativa | 11 | 500 | 12 | (Li et al., 2003) |
| Zea mays | 9 | 2400 | 10 | (Brady et al., 2007) |
| Gene 1 | Gene 2 | Ka | Ks | Ka/Ks | Duplication Date (Mya) |
|---|---|---|---|---|---|
| GmCOBL1 | GmCOBL14 | 0.051 | 0.104 | 0.491 | 8.55 |
| GmCOBL3 | GmCOBL7 | 0.026 | 0.112 | 0.233 | 9.17 |
| GmCOBL4 | GmCOBL9 | 0.021 | 0.157 | 0.131 | 12.84 |
| GmCOBL5 | GmCOBL8 | 0.019 | 0.092 | 0.204 | 7.55 |
| GmCOBL6 | GmCOBL19 | 0.224 | 0.134 | 1.677 | 10.97 |
| GmCOBL10 | GmCOBL18 | 0.017 | 0.139 | 0.124 | 11.38 |
| GmCOBL11 | GmCOBL21 | 0.110 | 0.190 | 0.579 | 9.02 |
| GmCOBL12 | GmCOBL22 | 0.008 | 0.089 | 0.090 | 7.30 |
| GmCOBL15 | GmCOBL17 | 0.036 | 0.172 | 0.210 | 14.08 |
| GmCOBL16 | GmCOBL23 | 0.031 | 0.140 | 0.224 | 11.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sangi, S.; Araújo, P.M.; Coelho, F.S.; Gazara, R.K.; Almeida-Silva, F.; Venancio, T.M.; Grativol, C. Genome-Wide Analysis of the COBRA-Like Gene Family Supports Gene Expansion through Whole-Genome Duplication in Soybean (Glycine max). Plants 2021, 10, 167. https://doi.org/10.3390/plants10010167
Sangi S, Araújo PM, Coelho FS, Gazara RK, Almeida-Silva F, Venancio TM, Grativol C. Genome-Wide Analysis of the COBRA-Like Gene Family Supports Gene Expansion through Whole-Genome Duplication in Soybean (Glycine max). Plants. 2021; 10(1):167. https://doi.org/10.3390/plants10010167
Chicago/Turabian StyleSangi, Sara, Paula M. Araújo, Fernanda S. Coelho, Rajesh K. Gazara, Fabrício Almeida-Silva, Thiago M. Venancio, and Clicia Grativol. 2021. "Genome-Wide Analysis of the COBRA-Like Gene Family Supports Gene Expansion through Whole-Genome Duplication in Soybean (Glycine max)" Plants 10, no. 1: 167. https://doi.org/10.3390/plants10010167
APA StyleSangi, S., Araújo, P. M., Coelho, F. S., Gazara, R. K., Almeida-Silva, F., Venancio, T. M., & Grativol, C. (2021). Genome-Wide Analysis of the COBRA-Like Gene Family Supports Gene Expansion through Whole-Genome Duplication in Soybean (Glycine max). Plants, 10(1), 167. https://doi.org/10.3390/plants10010167







