Targeted Metabolic and In-Silico Analyses Highlight Distinct Glucosinolates and Phenolics Signatures in Korean Rapeseed Cultivars
Abstract
:1. Introduction
2. Results
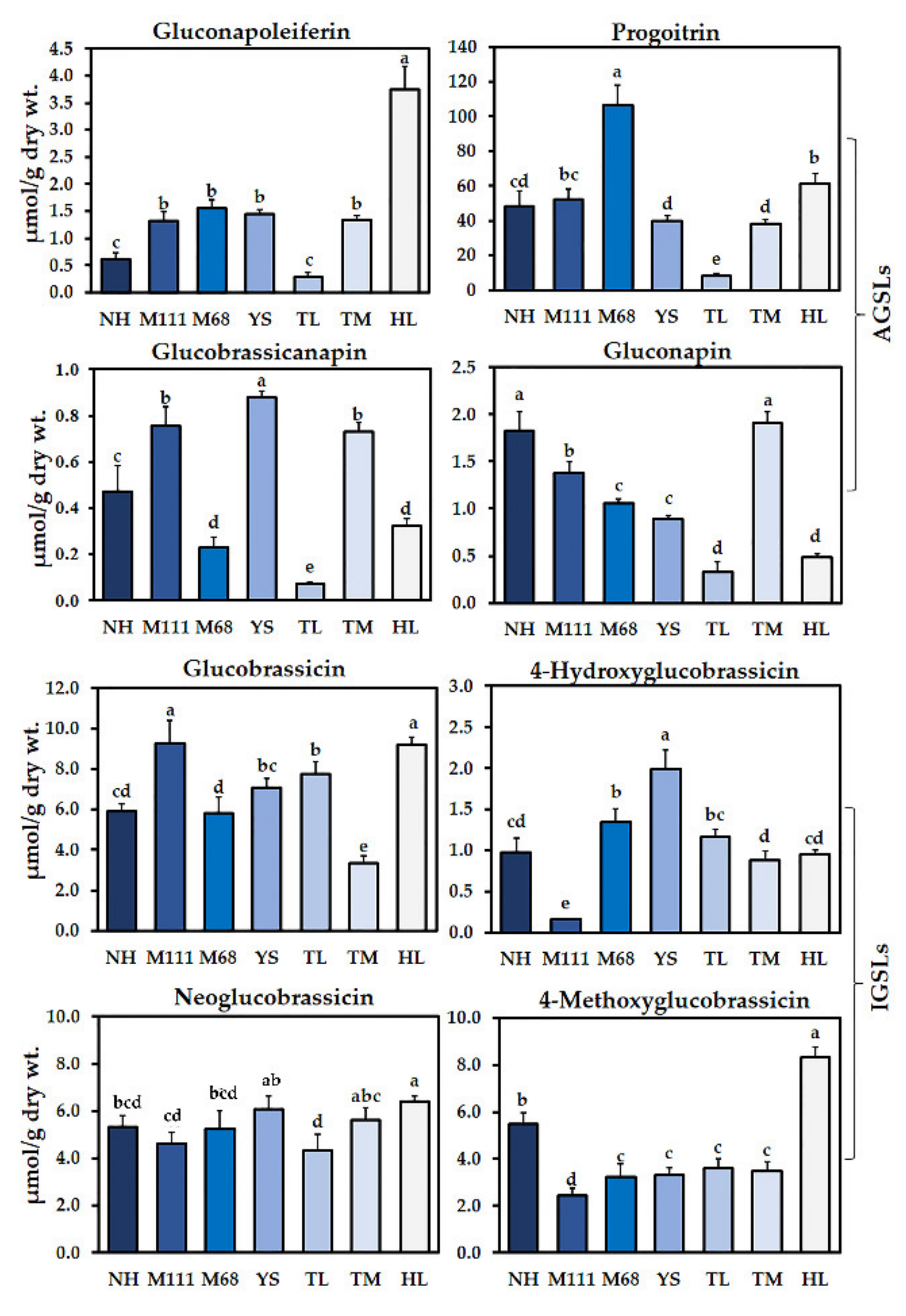
2.1. Examination of Glucosinolates (GSLs) in the Seven Korean Winter Cultivars of Rapeseed
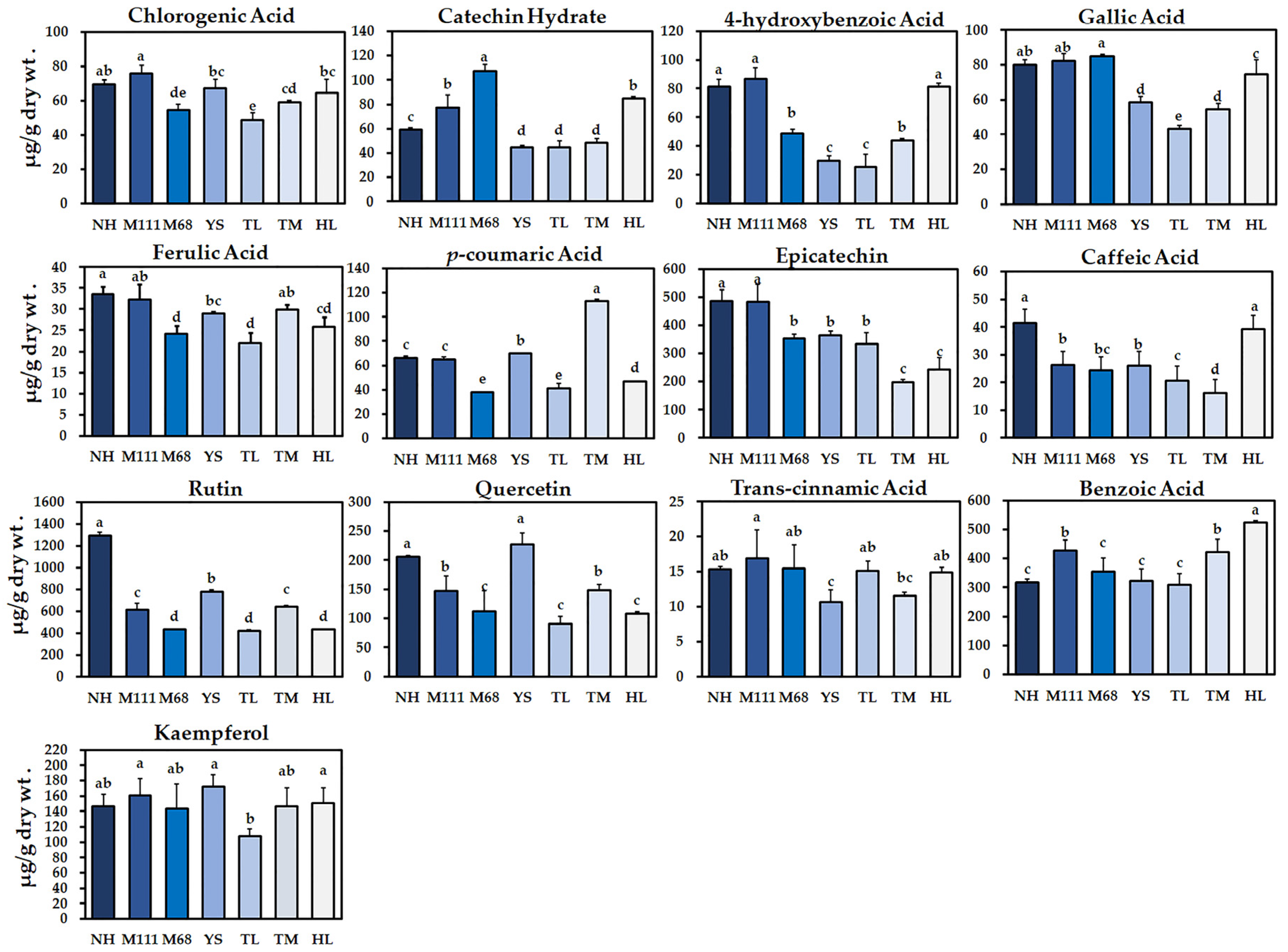
2.2. Determination of Phenolic Compounds in the Seven Korean Winter Cultivars of Rapeseed
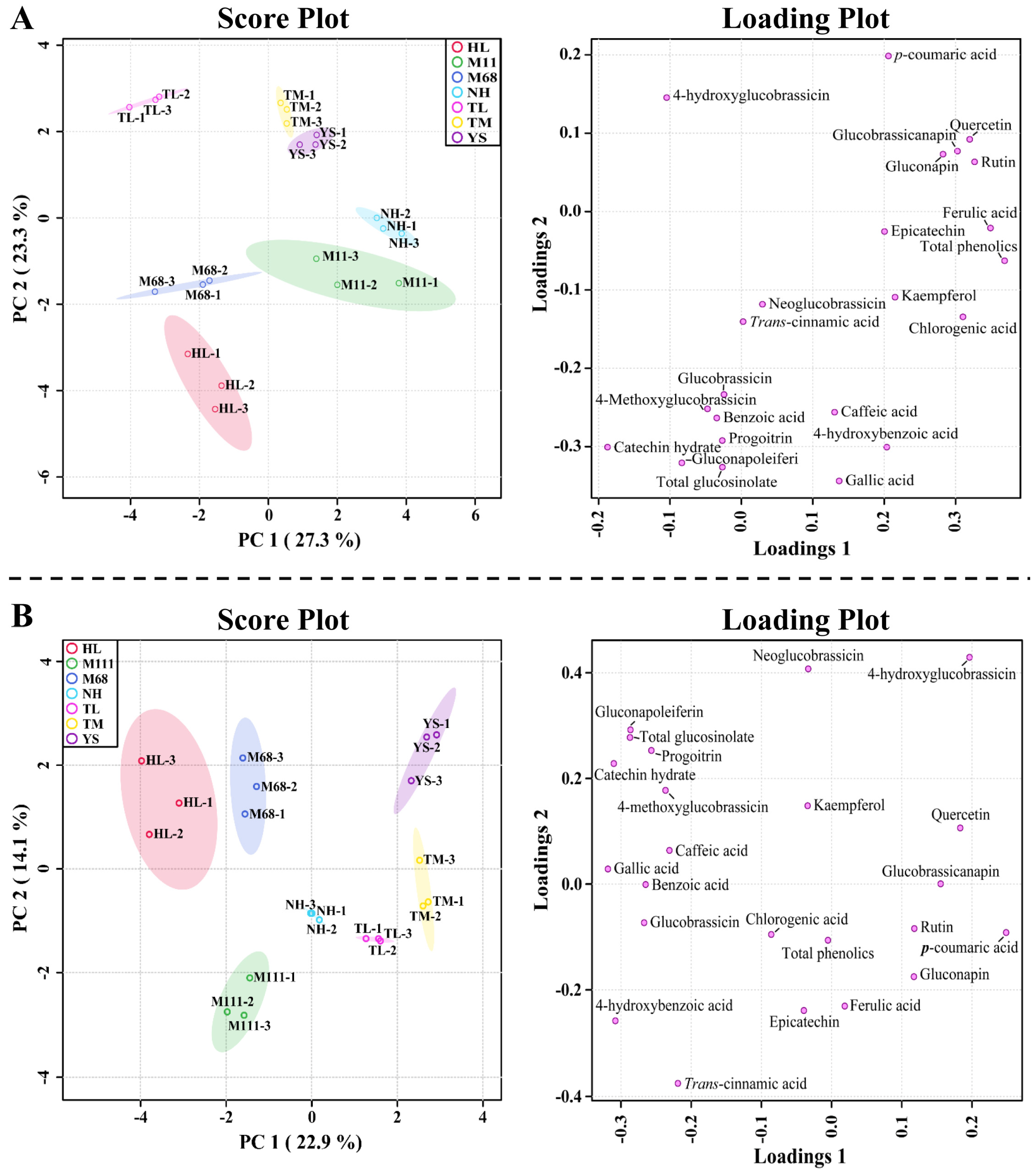
2.3. Relationships of GSLs and Phenolics in the Seven Korean Winter Rapeseed Cultivars
2.4. Metabolic Profiling of Identified Metabolites from Seven Korean Cultivars
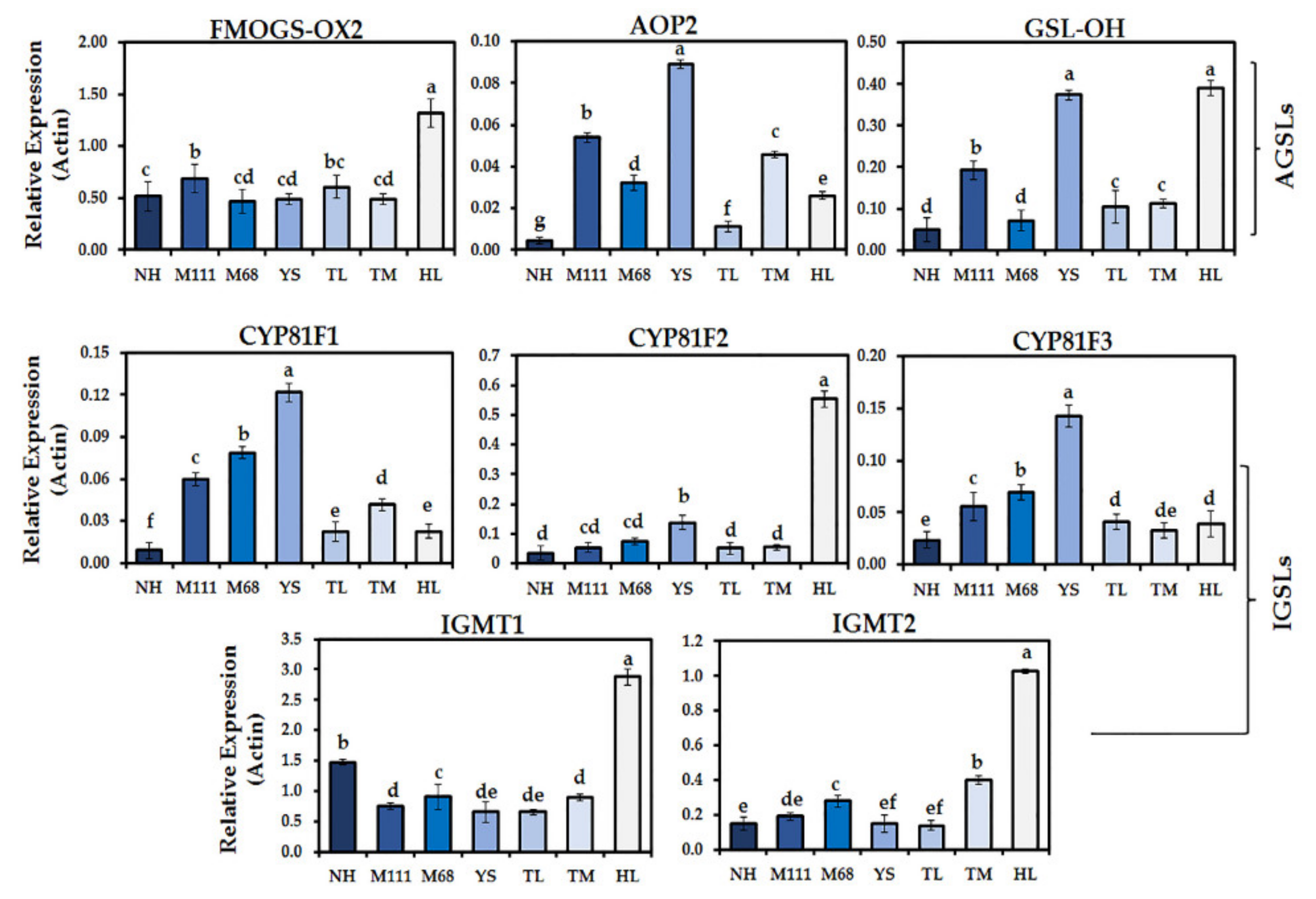
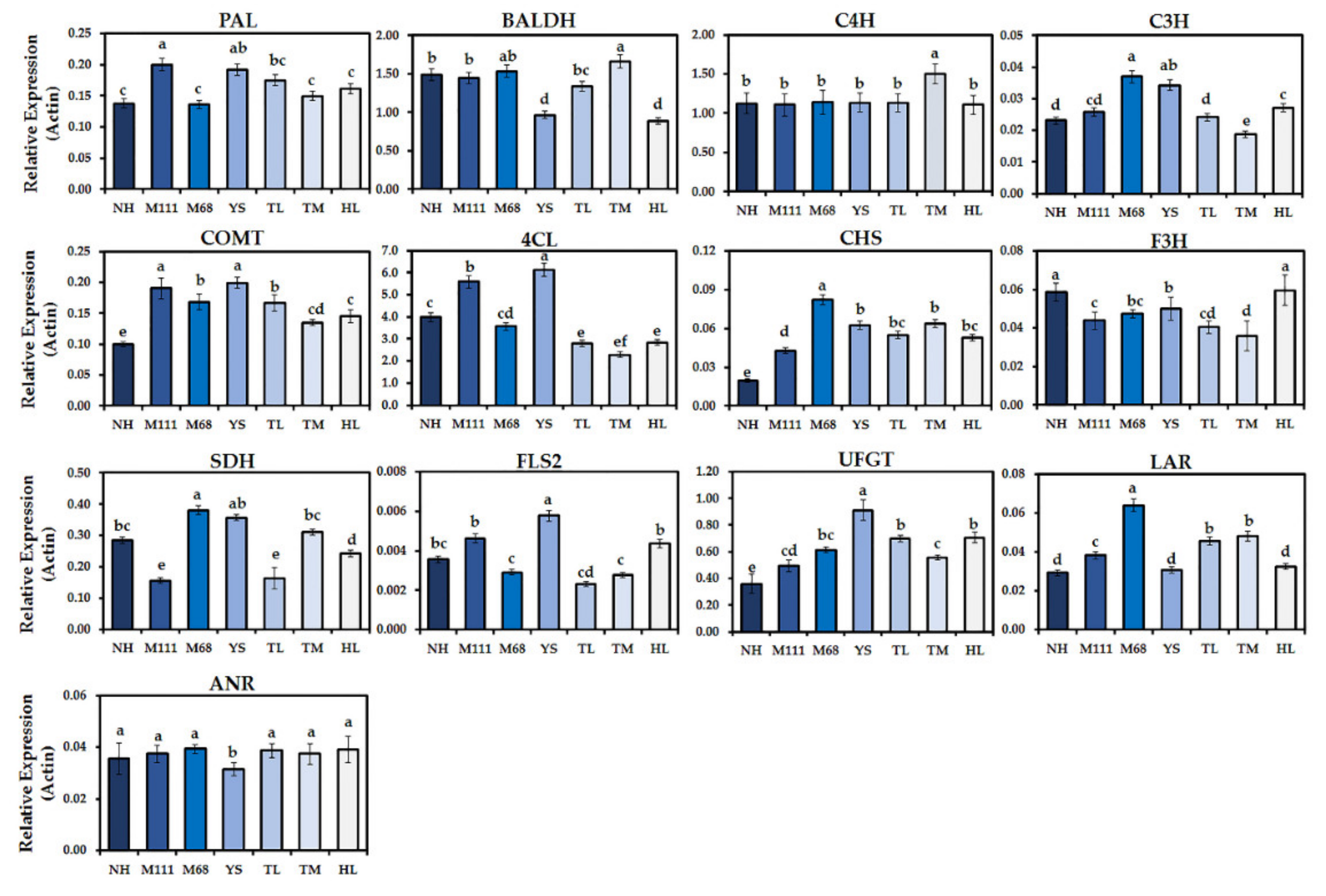
2.5. Gene Expression Analysis of Biosynthetic Pathways for GSLs and Phenolics
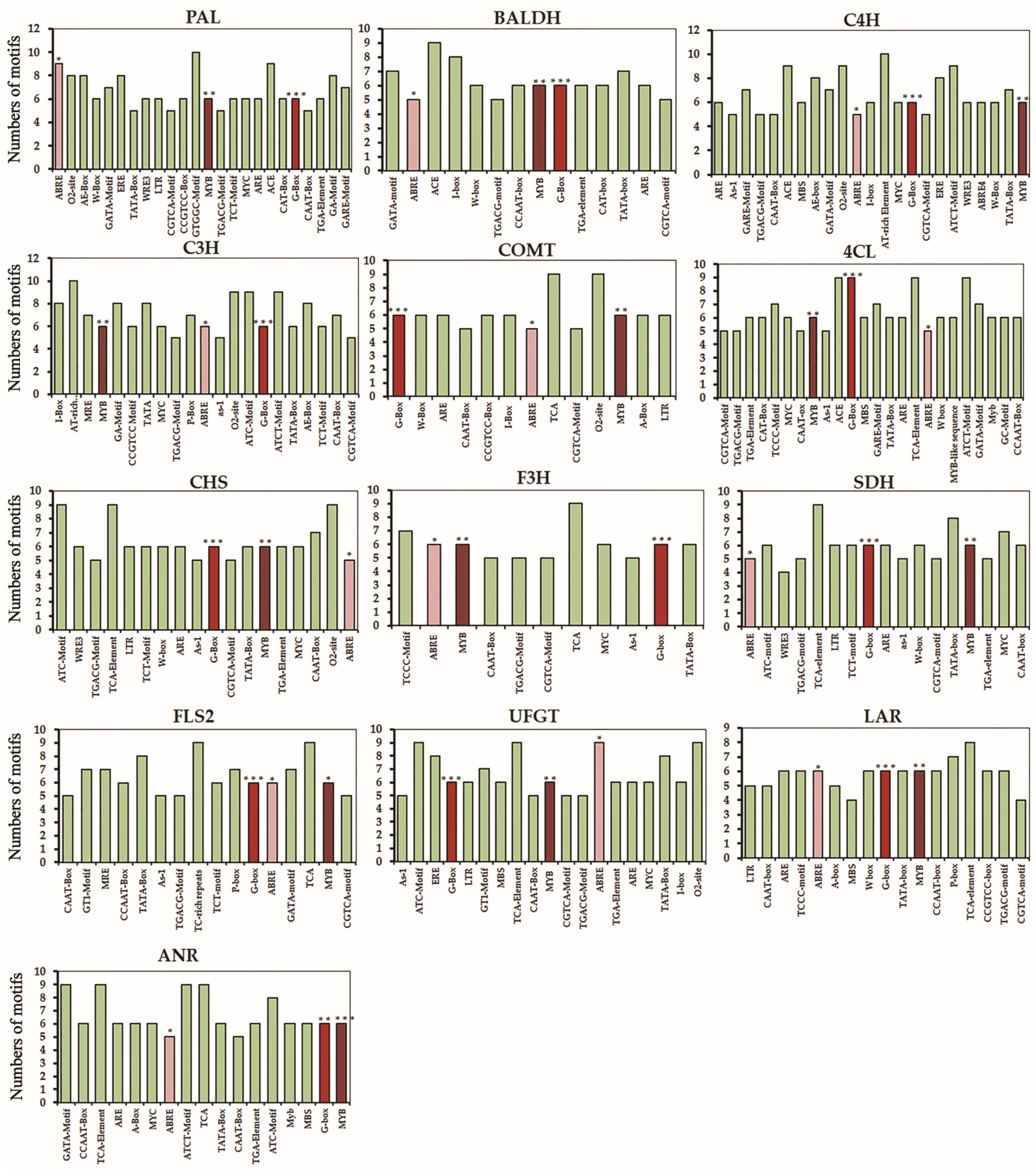
2.6. In Silico Analysis Highlights the cis-Regulatory Elements and Transcriptional Regulation Associated with Metabolite Changes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Extraction and Determination of Glucosinolates
4.3. Determination of Phenolic Compounds
4.4. RNA Extraction and Quantitative RT-PCR
4.5. Statistical and In Silico Analyses for the GSLs and Phenolics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bennouna, D.; Tourniaire, F.; Durand, T.; Galano, J.-M.; Fine, F.; Fraser, K.; Benatia, S.; Rosique, C.; Pau, C.; Couturier, C.; et al. The Brassica napus (oilseed rape) seeds bioactive health effects are modulated by agronomical traits as assessed by a mul-ti-scale omics approach in the metabolically impaired ob-mouse. Food Chem. Mol. Sci. 2021, 2, 100011. [Google Scholar] [CrossRef]
- Salehi, B.; Quispe, C.; Butnariu, M.; Sarac, I.; Marmouzi, I.; Kamle, M.; Tripathi, V.; Kumar, P.; Bouyahya, A.; Capanoglu, E.; et al. Phytotherapy and food applications from Brassica genus. Phytother. Res. 2021, 35, 3590–3609. [Google Scholar] [CrossRef]
- Jahangir, M.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Health-Affecting Compounds in Brassicaceae. Compr. Rev. Food Sci. Food Saf. 2009, 8, 31–43. [Google Scholar] [CrossRef]
- Yang, L.; Wen, K.-S.; Ruan, X.; Zhao, Y.-X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzin, V.; Galili, G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arab. Book 2010, 8, e0132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pott, D.M.; Osorio, S.; Vallarino, J.G. From Central to Specialized Metabolism: An Overview of Some Secondary Compounds Derived from the Primary Metabolism for Their Role in Conferring Nutritional and Organoleptic Characteristics to Fruit. Front. Plant Sci. 2019, 10, 835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangkadilok, N.; E Nicolas, M.; Bennett, R.N.; Premier, R.R.; Eagling, D.R.; Taylor, P. Developmental changes of sinigrin and glucoraphanin in three Brassica species (Brassica nigra, Brassica juncea and Brassica oleracea var. italica). Sci. Hortic. 2002, 96, 11–26. [Google Scholar] [CrossRef]
- Guerriero, G.; Berni, R.; Muñoz-Sánchez, J.A.; Apone, F.; Abdel-Salam, E.M.; Qahtan, A.A.; Alatar, A.A.; Cantini, C.; Cai, G.; Hausman, J.-F.; et al. Production of Plant Secondary Metabolites: Examples, Tips and Suggestions for Biotechnologists. Genes 2018, 9, 309. [Google Scholar] [CrossRef] [Green Version]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B. Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Cartea, M.E.; Francisco, M.; Soengas, P.; Velasco, P. Phenolic Compounds in Brassica Vegetables. Molecules 2010, 16, 251–280. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, G.; Peng, Y. A Critical Review on Phytochemical Profile and Biological Effects of Turnip (Brassica rapa L.). Front. Nutr. 2021, 8, 721733. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Rana, C. Plant secondary metabolites: A review. Int. J. Eng. Res. Gen. 2015, 3, 661–670. [Google Scholar]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- Plaszkó, T.; Szűcs, Z.; Vasas, G.; Gonda, S. Effects of Glucosinolate-Derived Isothiocyanates on Fungi: A Comprehensive Review on Direct Effects, Mechanisms, Structure-Activity Relationship Data and Possible Agricultural Applications. J. Fungi 2021, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, S.F. Glucosilates as natural pesticides. In Biologically Active Natural Products: Agrochemicals; CRC Press: Boca Raton, FL, USA, 1999; pp. 81–91. [Google Scholar]
- Ishida, M.; Hara, M.; Fukino, N.; Kakizaki, T.; Morimitsu, Y. Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 2014, 64, 48–59. [Google Scholar] [CrossRef] [Green Version]
- Herr, I.; Büchler, M.W. Dietary constituents of broccoli and other cruciferous vegetables: Implications for prevention and therapy of cancer. Cancer Treat. Rev. 2010, 36, 377–383. [Google Scholar] [CrossRef]
- Heber, D. Vegetables, fruits and phytoestrogens in the prevention of diseases. J. Postgrad. Med. 2004, 50, 145–149. [Google Scholar]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evi-dence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [Green Version]
- Shock, T.; Badang, L.; Ferguson, B.; Martinez-Guryn, K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J. Nutr. Biochem. 2021, 95, 108631. [Google Scholar] [CrossRef]
- Ernst, W. Ecological aspects of sulfur in higher plants: The impact of SO2 and the evolution of the biosynthesis of organic sul-fur compounds on populations and ecosystems. In Sulfur Nutrition and Sulfur Assimilation in Higher Plants: Regulatory, Agri-Cultural and Environmental Aspects; De Kok, L.J., Stulen, I., Rennenberg, H., Brunold, C., Rauser, W., Eds.; SPB Academic Publishing: The Hague, The Netherlands, 1993; pp. 295–313. [Google Scholar]
- Schnug, E. Physiological functions and environmental relevance of sulfur-containing secondary metabolites. In Sulfur Nutrition and Sulfur Assimilation in Higher Plant; Regulatory Agricultural and Environmental Aspects; De Kok, L.J., Stulen, I., Rennenberg, H., Brunold, C., Rauser, W., Eds.; SPB Academic Publishing: The Hague, The Netherlands, 1993; pp. 179–190. [Google Scholar]
- Del Carmen Martínez-Ballesta, M.; Moreno, D.A.; Carvajal, M. The physiological importance of glucosinolates on plant re-sponse to abiotic stress in Brassica. Int. J. Mol. Sci. 2013, 14, 11607–11625. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defence mechanisms. New Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Luo, Q.; Sun, M.; Corke, H. Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci. 2004, 74, 2157–2184. [Google Scholar] [CrossRef]
- Tiwari, U.; Cummins, E. Factors influencing levels of phytochemicals in selected fruit and vegetables during pre- and post-harvest food processing operations. Food Res. Int. 2013, 50, 497–506. [Google Scholar] [CrossRef]
- Ayaz, F.A.; Hayırlıoglu-Ayaz, S.; Alpay-Karaoglu, S.; Grúz, J.; Valentová, K.; Ulrichová, J.; Strnad, M. Phenolic acid contents of kale (Brassica oleraceae L. var. acephala DC.) extracts and their antioxidant and antibacterial activities. Food Chem. 2008, 107, 19–25. [Google Scholar] [CrossRef]
- Park, C.H.; Kim, N.S.; Park, J.S.; Lee, S.Y.; Lee, J.-W.; Park, S.U. Effects of light-emitting diodes on the accumulation of glu-cosinolates and phenolic compounds in sprouting canola (Brassica napus L.). Foods 2019, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Avato, P.; Argentieri, M.P. Brassicaceae: A rich source of health improving phytochemicals. Phytochem. Rev. 2015, 14, 1019–1033. [Google Scholar] [CrossRef]
- Björkman, M.; Klingen, I.; Birch, A.N.; Bones, A.M.; Bruce, T.J.; Johansen, T.J.; Meadow, R.; Mølmann, J.; Seljåsen, R.; Smart, L.E. Phytochemicals of Brassicaceae in plant protection and human health–Influences of climate, environment and agro-nomic practice. Phytochemistry 2011, 72, 538–556. [Google Scholar] [CrossRef]
- Wittkop, B.; Snowdon, R.J.; Friedt, W. Status and perspectives of breeding for enhanced yield and quality of oilseed crops for Europe. Euphytica 2009, 170, 131. [Google Scholar] [CrossRef]
- De Pascale, S.; Maggio, A.; Pernice, R.; Fogliano, V.; Barbieri, G. Sulphur fertilization may improve the nutritional value of Brassica rapa L. subsp. sylvestris. Eur. J. Agron. 2007, 26, 418–424. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Z.; Gerendás, J. Effects of Nitrogen and Sulfur on Total Phenolics and Antioxidant Activity in Two Genotypes of Leaf Mustard. J. Plant Nutr. 2008, 31, 1642–1655. [Google Scholar] [CrossRef]
- Brown, P.D.; Morra, M.J. Hydrolysis products of glucosinolates in Brassica napus tissues as inhibitors of seed germination. Plant Soil 1996, 181, 307–316. [Google Scholar] [CrossRef]
- Vierheilig, H.; Bennett, R.; Kiddle, G.; Kaldorf, M.; Ludwig-Müller, J. Differences in glucosinolate patterns and arbuscular mycorrhizal status of glucosinolate-containing plant species. New Phytol. 2000, 146, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasumoto, S.; Matsuzaki, M.; Hirokane, H.; Okada, K. Glucosinolate content in rapeseed in relation to suppression of subse-quent crop. Plant Prod. Sci. 2010, 13, 150–155. [Google Scholar] [CrossRef] [Green Version]
- Shahidi, F.; Gabon, J.E. Individual glucosinolates in six canola varieties. J. Food Qual. 1989, 11, 421–431. [Google Scholar] [CrossRef]
- Appelqvist, L.-A. Chemical constituents of rapeseed. In Rapeseed; Appelqvist, L., Ohlson, R., Eds.; Elsevier Publishing Co.: Amsterdam, The Netherlands, 1972; pp. 123–173. [Google Scholar]
- Jiang, J.; Zhang, C.; Wang, X. Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid in-sensitive 1. J. Integr. Plant Biol. 2013, 55, 1198–1211. [Google Scholar] [CrossRef] [Green Version]
- Auger, B.; Marnet, N.; Gautier, V.; Maia-Grondard, A.; Leprince, F.; Renard, M.; Guyot, S.; Nesi, N.; Routaboul, J.-M. A de-tailed survey of seed coat flavonoids in developing seeds of Brassica napus L. J. Agric. Food Chem. 2010, 58, 6246–6256. [Google Scholar] [CrossRef]
- Li, X.; Westcott, N.; Links, M.; Gruber, M.Y. Seed Coat Phenolics and the Developing Silique Transcriptome of Brassica carinata. J. Agric. Food Chem. 2010, 58, 10918–10928. [Google Scholar] [CrossRef]
- Marles, M.; Gruber, M.Y.; Scoles, G.J.; Muir, A.D. Pigmentation in the developing seed coat and seedling leaves of Brassica carinata is controlled at the dihydroflavonol reductase locus. Phytochemistry 2003, 62, 663–672. [Google Scholar] [CrossRef]
- Jiang, J.; Shao, Y.; Yanlin, S.; Lu, C.; Zhang, Y.; Wang, Y. Phenolic Composition Analysis and Gene Expression in Developing Seeds of Yellow- and Black-seeded Brassica napus. J. Integr. Plant Biol. 2013, 55, 537–551. [Google Scholar] [CrossRef]
- Chhajed, S.; Mostafa, I.; He, Y.; Abou-Hashem, M.; El-Domiaty, M.; Chen, S. Glucosinolate biosynthesis and the glucosin-olate–myrosinase system in plant defense. Agronomy 2020, 10, 1786. [Google Scholar] [CrossRef]
- Harun, S.; Abdullah-Zawawi, M.-R.; Goh, H.-H.; Mohamed-Hussein, Z.-A. A comprehensive gene inventory for glucosin-olate biosynthetic pathway in Arabidopsis thaliana. J. Agric. Food Chem. 2020, 68, 7281–7297. [Google Scholar] [CrossRef]
- Koornneef, M.; Meinke, D. The development of Arabidopsis as a model plant. Plant J. 2010, 61, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Wang, C.; Crocoll, C.; Halkier, B.A. Biotechnological approaches in glucosinolate production. J. Integr. Plant Biol. 2018, 60, 1231–1248. [Google Scholar] [CrossRef]
- Bell, L. The Biosynthesis of Glucosinolates: Insights, Inconsistencies, and Unknowns. Annu. Plant Rev. Online 2019, 2, 969–1000. [Google Scholar]
- Nguyen, V.P.T.; Stewart, J.; Lopez, M.; Ioannou, I.; Allais, F. Glucosinolates: Natural Occurrence, Biosynthesis, Accessibility, Isolation, Structures, and Biological Activities. Molecules 2020, 25, 4537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.; Huai, D.; Zhou, Y.; Kliebenstein, D.J. The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randhir, R.; Lin, Y.-T.; Shetty, K. Stimulation of phenolics, antioxidant and antimicrobial activities in dark germinated mung bean sprouts in response to peptide and phytochemical elicitors. Process. Biochem. 2004, 39, 637–646. [Google Scholar] [CrossRef]
- Lin, D.; Xiao, M.; Zhao, J.; Li, Z.; Xing, B.; Li, X.; Kong, M.; Li, L.; Zhang, Q.; Liu, Y. An overview of plant phenolic com-pounds and their importance in human nutrition and management of type 2 diabetes. Molecules 2016, 21, 1374. [Google Scholar] [CrossRef] [PubMed]
- Chhon, S.; Jeon, J.; Kim, J.; Park, S.U. Accumulation of Anthocyanins through Overexpression of AtPAP1 in Solanum nigrum Lin. (Black Nightshade). Biomolecules 2020, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, H.J.; Baek, S.-A.; Sathasivam, R.; Kim, J.K.; Park, S.U. Metabolomic analysis reveals the interaction of primary and sec-ondary metabolism in white, pale green, and green pak choi (Brassica rapa subsp. chinensis). Appl. Biol. Chem. 2021, 64, 1–16. [Google Scholar] [CrossRef]
- Kim, J.K.; Bong, S.J.; Park, S.U. Amino Acids Content in Different Korean Cultivars of Rapeseed (Brassica napus). Asian J. Chem. 2017, 29, 1423–1426. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; Morais, D.A.D.L.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Sohn, S.I.; Sathasivam, R.; Khaskheli, A.J.; Kim, M.C.; Kim, N.S.; Park, S.U. Targeted Metabolic and In-Silico Analyses Highlight Distinct Glucosinolates and Phenolics Signatures in Korean Rapeseed Cultivars. Plants 2021, 10, 2027. https://doi.org/10.3390/plants10102027
Kim J, Sohn SI, Sathasivam R, Khaskheli AJ, Kim MC, Kim NS, Park SU. Targeted Metabolic and In-Silico Analyses Highlight Distinct Glucosinolates and Phenolics Signatures in Korean Rapeseed Cultivars. Plants. 2021; 10(10):2027. https://doi.org/10.3390/plants10102027
Chicago/Turabian StyleKim, Joonyup, Soo In Sohn, Ramaraj Sathasivam, Allah Jurio Khaskheli, Min Cheol Kim, Nam Su Kim, and Sang Un Park. 2021. "Targeted Metabolic and In-Silico Analyses Highlight Distinct Glucosinolates and Phenolics Signatures in Korean Rapeseed Cultivars" Plants 10, no. 10: 2027. https://doi.org/10.3390/plants10102027
APA StyleKim, J., Sohn, S. I., Sathasivam, R., Khaskheli, A. J., Kim, M. C., Kim, N. S., & Park, S. U. (2021). Targeted Metabolic and In-Silico Analyses Highlight Distinct Glucosinolates and Phenolics Signatures in Korean Rapeseed Cultivars. Plants, 10(10), 2027. https://doi.org/10.3390/plants10102027







