Genetic Stability, Phenolic, Flavonoid, Ferulic Acid Contents, and Antioxidant Activity of Micropropagated Lycium schweinfurthii Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Culture Conditions
2.2. In Vitro Micropropagation
2.3. DNA Extraction and PCR Amplification Conditions
2.4. Protein Extraction and SDS-PAGE
2.5. Secondary Metabolites
2.5.1. Sample Preparation and Extraction
2.5.2. Total Phenolic Assay
2.5.3. Total Flavonoid Assay
2.5.4. HPTLC Conditions
2.6. Antioxidant Capacity
2.6.1. DPPH Free Radical Scavenging
2.6.2. ABTS Free Radical Scavenging
2.7. Recording Data and Statistical Analysis
3. Results and Discussion
3.1. In Vitro Propagation
3.2. Genetic Stability of Micropropagated Plantlets
3.3. Phenolic and Flavonoid Content Estimation
3.4. HPTLC Analysis
3.5. Antioxidant Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yao, R.; Heinrich, M.; Weckerle, C.S. The Genus Lycium as Food and Medicine: A Botanical, Ethnobotanical and Historical Review. J. Ethnopharmacol. 2018, 212, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Stearn, W.T. Lycium. In Flora Europaea; Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Webb, D.A., Eds.; Cambridge University Press: Cambridge, UK, 1972; p. 194. [Google Scholar]
- Shaltout, K.H.; El-Din, A.S.; El-Fahar, R.A.; Beshara, H.M. Associated Species and Threats upon Lycium Schweinfurthii var. Schweinfurthii in the Deltaic Mediterranean Coast, Egypt. Taeckholmia 2018, 38, 107–122. [Google Scholar]
- El-Amier, Y.; El-Halawany, E.; Abdullah, T. Composition and Diversity of Plant Communities in Sand Formations Along the Northern Coast of the Nile Delta in Egypt. Res. J. Pharm. Biol. Chem. Sci. 2014, 5, 826–847. [Google Scholar]
- Khafagi, A.; El- Ghamery, A.; Ghaly, O.; Ragab, O. Fruit and Seed Morphology of Some Species of Solanaceae. Taeckholmia 2018, 38, 123–140. [Google Scholar] [CrossRef]
- Cardoso, J.C.; Sheng Gerald, L.T.; Teixeira da Silva, J.A. Micropropagation in the Twenty-First Century. Methods Mol. Biol. 2018, 1815, 17–46. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Deng, Y.; Zheng, K.; Hu, X.; Zhu, M.; Deng, X.; Xi, R. An Efficient Micropropagation Protocol for an Endangered Ornamental Tree Species (Magnolia sirindhorniae Noot. & Chalermglin) and Assessment of Genetic Uniformity through DNA Markers. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Kikowska, M.; Thiem, B.; Szopa, A.; Ekiert, H. Accumulation of Valuable Secondary Metabolites: Phenolic Acids and Flavonoids in Different in vitro Systems of Shoot Cultures of the Endangered Plant Species—Eryngium alpinum L. Plant Cell Tissue Organ Cult. 2020, 141, 381–391. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Q.-M.; An, Q.; Cui, J.; Zhou, Y.; Qi, X.; Zhang, L.; Li, L. A Novel Micropropagation of Lycium ruthenicum and Epigenetic Fidelity Assessment of Three Types of Micropropagated Plants in vitro and ex vitro. PLoS ONE 2021, 16, e0247666. [Google Scholar] [CrossRef]
- Samiei, L.; Mirshahi, H.; Pahnehkolayi, M.D.; Tehranifar, A. Micropropagation of Lycium depressum, a Promising Native Shrub for Urban landscape. Iran. J. Hortic. Sci. 2020, 51, 741–751. [Google Scholar] [CrossRef]
- Silvestri, C.; Sabbatini, G.; Marangelli, F.; Rugini, E.; Cristofori, V. Micropropagation and ex vitro Rooting of Wolfberry. HortScience 2018, 53, 1494–1499. [Google Scholar] [CrossRef]
- Jung, W.-S.; Chung, I.-M.; Kim, S.-H.; Chi, H.-Y.; Yu, C.Y.; Ghimire, B.K. Direct Shoot Organogenesis from Lycium chinense Miller Leaf Explants and Assessment of Genetic Stability Using ISSR Markers. Agronomy 2021, 11, 503. [Google Scholar] [CrossRef]
- Samaha, G.M.; Ahmed, M.A.; Abd El-Hameid, A.R. Assessment of Growth and Productivity of Five Peanut Cultivars and Genetic Diversity Using RAPD Markers. Bull. Natl. Res. Cent. 2019, 43, 1–11. [Google Scholar] [CrossRef]
- Thakur, M.; Soni, M.; Sharma, D.P.; Vivek, M.; Sharma, V. In vitro Propagation of Plum (Prunus salicina) cv. ‘Santa Rosa’ and Assessment of Genetic Stability Using RAPD Markers. Indian J. Plant Physiol. 2018, 23, 161–168. [Google Scholar] [CrossRef]
- Lema-Rumińska, J.; Kulus, D.; Tymoszuk, A.; Varejão, J.M.T.B.; Bahcevandziev, K. Profile of Secondary Metabolites and Genetic Stability Analysis in New Lines of Echinacea purpurea (L.) Moench Micropropagated via Somatic Embryogenesis. Ind. Crops Prod. 2019, 142, 111851. [Google Scholar] [CrossRef]
- Tikendra, L.; Potshangbam, A.M.; Dey, A.; Devi, T.R.; Sahoo, M.R.; Nongdam, P. RAPD, ISSR, and SCoT Markers Based Genetic Stability Assessment of Micropropagated Dendrobium fimbriatum Lindl. Var. Oculatum Hk. f.-an Important Endangered Orchid. Physiol. Mol. Biol. Plants 2021, 27, 341–357. [Google Scholar] [CrossRef]
- Oliya, B.K.; Chand, K.; Thakuri, L.S.; Baniya, M.K.; Sah, A.K.; Pant, B. Assessment of Genetic Stability of Micropropagated Plants of Rhynchostylis retusa (L.) Using RAPD Markers. Sci. Hortic. 2021, 281, 110008. [Google Scholar] [CrossRef]
- Patel, P.; Rajkumar, B.K.; Parmar, P.; Shah, R.; Krishnamurthy, R. Assessment of Genetic Diversity in Colletotrichum falcatum Went Accessions Based on RAPD and ISSR Markers. J. Genet. Eng. Bio-Technol. 2018, 16, 153–159. [Google Scholar] [CrossRef]
- Wójcik, D.; Trzewik, A.; Kucharska, D. Field Performance and Genetic Stability of Micropropagated Gooseberry Plants (Ribes grossularia L.). Agronomy 2021, 11, 45. [Google Scholar] [CrossRef]
- Tikendra, L.; Amom, T.; Nongdam, P. Molecular Genetic Homogeneity Assessment of Micropropagated Dendrobium moschatum Sw.—A Rare Medicinal Orchid, Using RAPD and ISSR Markers. Plant Gene 2019, 19, 100196. [Google Scholar] [CrossRef]
- Chittora, M. Assessment of Genetic Fidelity of Long Term Micropropagated Shoot Cultures of Achras sapota L. Var. “Cricket Ball” as Assessed by RAPD and ISSR Markers. Indian J. Biotechnol. 2018, 17, 492–495. [Google Scholar]
- Ahmed, M.R.; Anis, M.; Alatar, A.A.; Faisal, M. In Vitro Clonal Propagation and Evaluation of Genetic Fidelity Using RAPD and ISSR Marker in Micropropagated Plants of Cassia alata L.: A Potential Medicinal Plant. Agrofor. Syst. 2017, 91, 637–647. [Google Scholar] [CrossRef]
- Jong, L.W.; Thien, V.Y.; Yong, Y.S.; Rodrigues, K.F.; Yong, W.T.L. Micropropagation and Protein Profile Analysis by SDS-PAGE of Gracilaria changii (Rhodophyta, Solieriaceae). Aquac. Rep. 2015, 1, 10–14. [Google Scholar] [CrossRef]
- Khattab, S.; El Sherif, F.; El-Garhy, H.A.; Ahmed, S.; Ibrahim, A. Genetic and Phytochemical Analysis of the in vitro Regenerated Pilosocereus robinii by ISSR, SDS-PAGE and HPLC. Gene 2014, 533, 313–321. [Google Scholar] [CrossRef]
- Mahmoud, R.A.; Hassan, O.S.; Abou-Hashish, A.; Amin, A. Role of Trehalose during Recovery from Drought Stress in Micropropagated Banana (Musa Spp.) Transplants. Res. J. Pharm. Biol. Chem. Sci. 2017, 8, 1335–1345. [Google Scholar]
- El-Mageid, I.S. Evaluation of Genetic Stability by Using Protein and ISSR Markers during Callus Development Stage of Some Date Palm (Phoenix dactylifera L.) Cultivars under Effect of 2,4-D and Picloram. Middle East J. Appl. Sci. 2019, 9, 483–493. [Google Scholar]
- Konarska, A. Microstructural and Histochemical Characteristics of Lycium barbarum L. Fruits Used in Folk Herbal Medicine and as Functional Food. Protoplasma 2018, 255, 1839–1854. [Google Scholar] [CrossRef]
- Wink, M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef] [PubMed]
- Ewais, E.A.; Abd El-Maboud, M.M.; Elhaw, M.H.; Haggag, M.I. Phytochemical Studies on Lycium schweinfurthii Var. Schweinfurthii (Solanaceae) and Isolation of Five Flavonoids from Leaves. J. Med. Plants Stud. 2016, 4, 288–300. [Google Scholar]
- Cushnie, T.P.T.; Lamb, A.J. Recent Advances in Understanding the Antibacterial Properties of Flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef]
- Tattini, M.; Galardi, C.; Pinelli, P.; Massai, R.; Remorini, D.; Agati, G. Differential Accumulation of Flavonoids and Hydroxycinnamates in Leaves of Ligustrum vulgare under Excess Light and Drought Stress. New Phytol. 2004, 163, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Barceló, J.; Poschenrieder, C. Fast Root Growth Responses, Root Exudates, and Internal Detoxification as Clues to the Mechanisms of Aluminium Toxicity and Resistance: A Review. Environ. Exp. Bot. 2002, 48, 75–92. [Google Scholar] [CrossRef]
- Ryan, K.G.; Swinny, E.E.; Markham, K.R.; Winefield, C. Flavonoid Gene Expression and UV Photoprotection in Transgenic and Mutant Petunia Leaves. Phytochemistry 2002, 59, 23–32. [Google Scholar] [CrossRef]
- Treutter, D. Significance of Flavonoids in Plant Resistance: A Review. Environ. Chem. Lett. 2006, 4, 147–157. [Google Scholar] [CrossRef]
- Lin, D.; Xiao, M.; Zhao, J.; Li, Z.; Xing, B.; Li, X.; Kong, M.; Li, L.; Zhang, Q.; Liu, Y.; et al. An Overview of Plant Phenolic Compounds and Their Importance in Human Nutrition and Management of Type 2 Diabetes. Molecules 2016, 21, 1374. [Google Scholar] [CrossRef]
- Pietta, P.G. Flavonoids as Antioxidants. J. Nat. Prod. 2000, 63, 1035–1042. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-Inflammatory Effects of Flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Gaynor, R.B. Therapeutic Potential of Inhibition of the NF-κB Pathway in the Treatment of Inflammation and Cancer. J. Clin. Invest. 2001, 107, 135–142. [Google Scholar] [CrossRef]
- Lani, R.; Hassandarvish, P.; Shu, M.H.; Phoon, W.H.; Chu, J.J.H.; Higgs, S.; Vanlandingham, D.; Abu Bakar, S.; Zandi, K. Antiviral Activity of Selected Flavonoids against Chikungunya Virus. Antiviral Res. 2016, 133, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Manner, S.; Skogman, M.; Goeres, D.; Vuorela, P.; Fallarero, A. Systematic Exploration of Natural and Synthetic Flavonoids for the Inhibition of Staphylococcus aureus Biofilms. Int. J. Mol. Sci. 2013, 14, 19434–19451. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.J.; Dwyer, J.T.; Jacques, P.F.; McCullough, M.L. Associations between Flavonoids and Cardiovascular Disease Incidence or Mortality in European and US Populations. Nutr. Rev. 2012, 70, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Rezende, B.A.; Pereira, A.; Cortes, S.; Lemos, V. Vascular Effects of Flavonoids. Curr. Med. Chem. 2016, 23, 87–102. [Google Scholar] [CrossRef]
- Cazarolli, L.; Zanatta, L.; Alberton, E.; Bonorino Figueiredo, M.S.; Folador, P.; Damazio, R.; Pizzo-latti, M.; Barreto Silva, F.R. Flavonoids: Prospective Drug Candidates. Mini-Rev. Med. Chem. 2008, 8, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Jamous, R.; Zaitoun, S.; Husein, A.; Qasem, I.; Ali-Shtayeh, M. Screening for Biological Activities of Medicinal Plants Used in Traditional Arabic Palestinian Herbal Medicine. Eur. J. Med. Plants 2015, 9, 1–13. [Google Scholar] [CrossRef]
- Elbermawi, A.; Halim, A.F.; Mansour, E.S.S.; Ahmad, K.F.; Ashour, A.; Amen, Y.; Shimizu, K. A New Glucoside with a Potent α-Glucosidase Inhibitory Activity from Lycium schweinfurthii. Nat. Prod. Res. 2021, 35, 976–983. [Google Scholar] [CrossRef]
- Smetanska, I. Sustainable Production of Polyphenols and Antioxidants by Plant in vitro Cultures. In Bioprocessing of Plant In Vitro Systems; Pavlov, A., Bley, T., Eds.; Springer: Cham, Switzerland, 2018; pp. 225–269. ISBN 978-3-319-54599-8. [Google Scholar]
- Ali, A.M.A.; El-Nour, M.E.A.M.; Yagi, S.M. Total Phenolic and Flavonoid Contents and Antioxidant Activity of Ginger (Zingiber officinale Rosc.) Rhizome, Callus and Callus Treated with Some Elicitors. J. Genet. Eng. Biotechnol. 2018, 16, 677–682. [Google Scholar] [CrossRef]
- El-Hawary, S.S.; Abd El-Kader, E.M.; Rabeh, M.A.; Abdel Jaleel, G.A.; Arafat, M.A.; Schirmeister, T.; Abdelmohsen, U.R. Eliciting Callus Culture for Production of Hepatoprotective Flavonoids and Phenolics from Sequoia sempervirens (D. Don Endl). Nat. Prod. Res. 2020, 34, 3125–3129. [Google Scholar] [CrossRef]
- Darwish, H.Y.; Ahmed, S.M. Elicitors Enhancing Phenolics Content and Related Gene Expression Variation in Petal—Derived Calli of Rosa damascena Mill. Egypt. J. Bot. 2020, 60, 71–79. [Google Scholar] [CrossRef]
- Çelik, M.; Keskin, N.; Özdemir, F.A. The Effects of UV Irradiation and Incubation Time on in vitro Phenolic Compound Production in “Karaerik” Callus Culture. Kahramanmaraş Sütçü İmam Üniversitesi Tarım Ve Doğa Derg. 2020, 23, 1428–1434. [Google Scholar] [CrossRef]
- Martins, M.; Tenreiro, R.; Oliveira, M.M. Genetic Relatedness of Portuguese Almond Cultivars Assessed by RAPD and ISSR Markers. Plant Cell Rep. 2003, 22, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA Polymorphisms Amplified by Arbitrary Primers Are Useful as Genetic Markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Marinova, D.; Ribarova, F.; Atanassova, M. Total Phenolics and Total Flavonoids in Bulgarian Fruits and Vegetables. J. Univ. Chem. Technol. Metall. 2005, 40, 255–260. [Google Scholar]
- Olalere, O.A.; Abdurahman, H.N.; Yunus, R.B.M.; Alara, O.R.; Ahmad, M.M.; Zaki, Y.H.; Abdlrhman, H.S.M. Parameter Study, Antioxidant Activities, Morphological and Functional Characteristics in Microwave Extraction of Medicinal Oleoresins from Black and White Pepper. J. Taibah Univ. Sci. 2018, 12, 730–737. [Google Scholar] [CrossRef]
- Gabr, A.M.M.; Mabrok, H.B.; Ghanem, K.Z.; Blaut, M.; Smetanska, I. Lignan Accumulation in Callus and Agrobacterium Rhizogenes-Mediated Hairy Root Cultures of Flax (Linum usitatissimum). Plant Cell Tissue Organ Cult. 2016, 126, 255–267. [Google Scholar] [CrossRef]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; W. H. Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Hasan, M.N.; Nigar, S.; Rabbi, M.A.K.; Mizan, S.B.; Rahman, M.S. Micropropagation Of Strawberry (Fragaria x Ananassa Duch.). Int. J. Sustain. Crop. Prod. 2010, 5, 36–41. [Google Scholar]
- Amghar, I.; Diria, G.; Boumlik, I.; Gaboun, F.; Iraqi, D.; Labhilili, M.; Mentag, R.; Meziani, R.; Mazri, M.A.; Ibriz, M.; et al. An Efficient Regeneration Pathway through Adventitious Organogenesis for the Endangered Argania spinosa (L.) Skeels. Vegetos 2021, 34, 355–367. [Google Scholar] [CrossRef]
- Thomas, T.D.; Maseena, E.A. Callus Induction and Plant Regeneration in Cardiospermum halicacabum Linn. an Important Medicinal Plant. Sci. Hortic. 2006, 108, 332–336. [Google Scholar] [CrossRef]
- Irvani, N.; Solouki, M.; Omidi, M.; Zare, A.R.; Shahnazi, S. Callus Induction and Plant Regeneration in Dorem ammoniacum D., an Endangered Medicinal Plant. Plant Cell Tissue Organ Cult. 2010, 100, 293–299. [Google Scholar] [CrossRef]
- Sen, M.K.; Nasrin, S.; Rahman, S.; Jamal, A.H.M. In Vitro Callus Induction and Plantlet Regeneration of Achyranthes aspera L., a High Value Medicinal Plant. Asian Pac. J. Trop. Biomed. 2014, 4, 40–46. [Google Scholar] [CrossRef]
- Ozdemir, F.A.; Gur, N. In Vitro Propagation of Cataloglu Apricot (Prunus armeniaca L.) Cultivar Using Apical Node As Explant. Prog. Nutr. 2018, 20, 176–181. [Google Scholar] [CrossRef]
- El-Ghamery, A.A.; Khafagi, A.A.F.; Ragab, O.G. Taxonomic Implication of Pollen Morphology and Seed Protein Electrophoresis of Some Species of Solanaceae in Egypt. Azhar Bull. Sci. 2018, 29, 43–54. [Google Scholar]
- Khawale, R.N.; Singh, S.K.; Yerramilli, V.; Grover, M. Assessment of Clonal Fidelity of Micropropagated Grape (Vitis vinifera L.) Plants by RAPD Analysis. Physiol. Mol. Biol. Plants 2006, 12, 189–192. [Google Scholar]
- Lakshmanan, V.; Reddampalli Venkataramareddy, S.; Neelwarne, B. Molecular Analysis of Genetic Stability in Long-Term Micropropagated Shoots of Banana Using RAPD and ISSR Markers. Electron. J. Biotechnol. 2007, 10, 717–3458. [Google Scholar] [CrossRef]
- Martins, M.; Sarmento, D.; Oliveira, M.M. Genetic Stability of Micropropagated Almond Plantlets, as Assessed by RAPD and ISSR Markers. Plant Cell Rep. 2004, 23, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Parida, R.; Mohanty, S.; Nayak, S. Molecular Characterization of Endangered Medicinal Plant Species Hedychium coronarium from Eastern India. Int. J. Pharm. Pharm. Sci. 2017, 9, 173–178. [Google Scholar] [CrossRef]
- Osman, G.; Munshi, A.; Altf, F.; Mutawie, H. Genetic Variation and Relationships of Zea mays and Sorghum Species Using RAPD-PCR and SDS-PAGE of Seed Proteins. Afr. J. Biotechnol. 2013, 12, 4269–4276. [Google Scholar] [CrossRef][Green Version]
- Seesangboon, A.; Pokawattana, T.; Eungwanichayapant, P.D.; Tovaranonte, J.; Popluechai, S. Effects of 6-Benzyladenine on Jatropha Gene Expression and Flower Development. Russ. J. Plant Physiol. 2018, 65, 345–356. [Google Scholar] [CrossRef]
- Alizadeh, M.; Singh, S.K. Molecular Assessment of Clonal Fidelity in Micropropagated Grape (Vitis Spp.) Rootstock Genotypes Using RAPD and ISSR Markers. Iran. J. Biotechnol. 2009, 7, 17–44. [Google Scholar]
- Sulaiman, C.T.; Balachandran, I. Total phenolics and total flavonoids in selected Indian medicinal plants. Indian J. Pharm. Sci. 2012, 74, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Sultana, B.; Anwar, F. LC–ESI–MS/MS Based Characterization of Phenolic Components in Fruits of Two Species of Solanaceae. J. Food Sci. Technol. 2018, 55, 2370–2376. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Singh, A.P.; Singh Rawat, A.K. A HPTLC Method for the Identification of Ferulic Acid from Lycopodium clavatum. Asian Pac. J. Trop. Biomed. 2012, 2, S12–S14. [Google Scholar] [CrossRef]
- Goudar, G.; Sathisha, G.J. Effect of Processing on Ferulic Acid Content in Foxtail Millet (Setaria italica) Grain Cultivars Evaluated by HPTLC. Orient. J. Chem. 2016, 32, 2251–2258. [Google Scholar] [CrossRef]
- Verma, S.C.; Rani, R.; Pant, P.; Padhi, M.M.; Jain, C.L.; Babu, R. Quantitative Determination of Ferulic Acid in Ricinus communis Linn. Leaves and Its Geographical Variation Using HPTLC Finger-print. Chem. Sin. 2011, 2, 127–135. [Google Scholar]


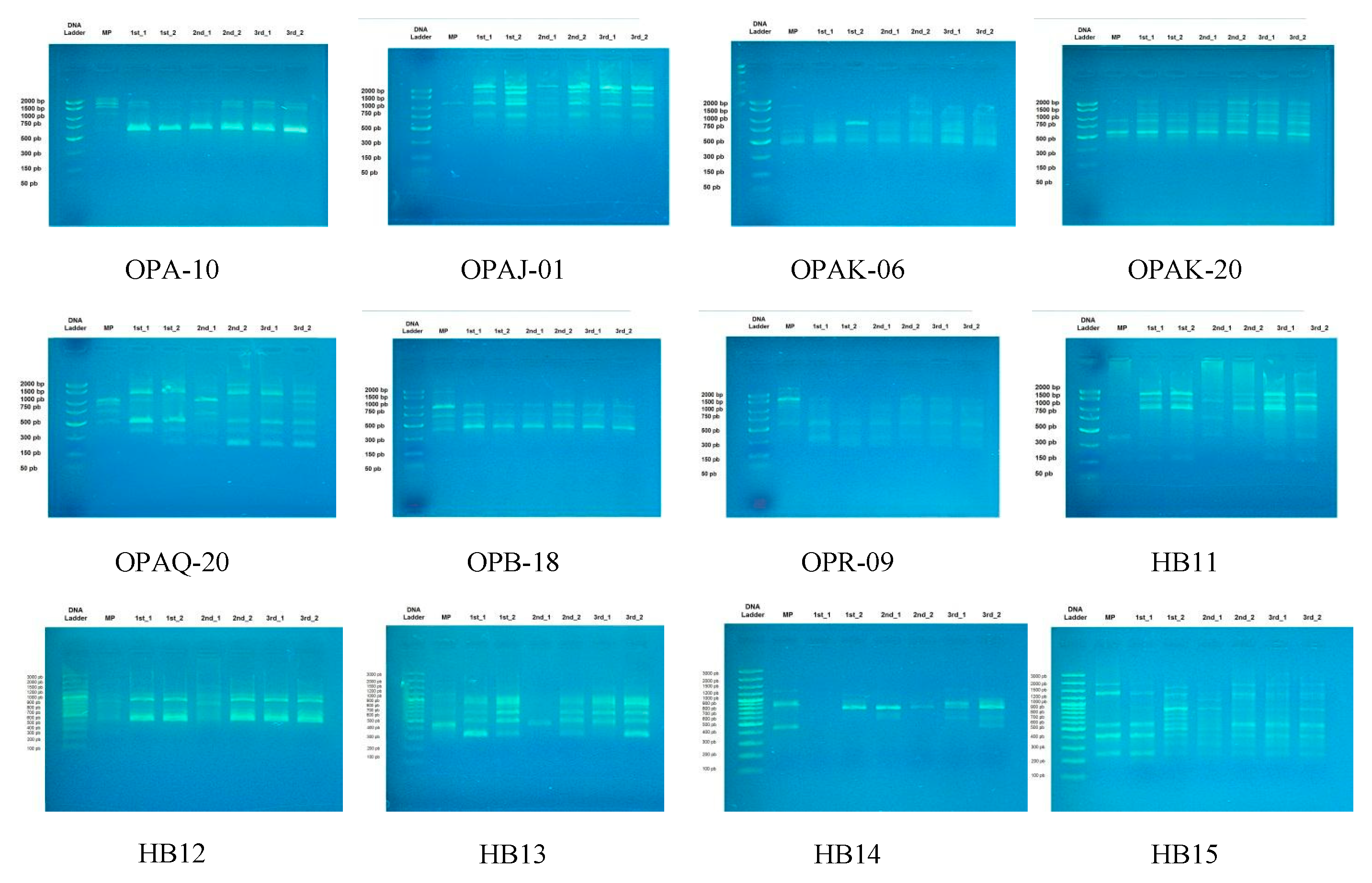



| Primer Name | Sequence | Annealing Temperature (°C) | |
|---|---|---|---|
| RAPD primers | OPA-10 | 5′-GTGATCGCAG-3′ | 40.5 |
| OPAJ-01 | 5′-ACGGGTCAGA-3′ | 43 | |
| OPAK-06 | 5′-TCACGTCCCT-3′ | 42 | |
| OPAK-20 | 5′-TGATGGCGTC-3′ | 41 | |
| OPAQ-20 | 5′-GTGAACGCTC-3′ | 40.5 | |
| OPB-18 | 5′-CCACAGCAGT-3′ | 42 | |
| OPR-09 | 5′-TGAGCACGAG-3′ | 42 | |
| ISSR primers | HB11 | 5′-GTGTGTGTGTGTCC-3′ | 54 |
| HB12 | 5′-CACCACCACGC-3′ | 50.9 | |
| HB13 | 5′- GAGGAGGAGGC-3′ | 48 | |
| HB14 | 5′-CTCCTCCTCGC-3′ | 48 | |
| HB15 | 5′-GTGGTGGTGGC-3′ | 50.9 | |
| Treatment (mg L−1) | Number of Plantlets | Number of Distinct Nodes | Number of Leaves | Shootlet Length (cm) | ||
|---|---|---|---|---|---|---|
| BA | Kin | NAA | ||||
| - | - | - | 1.00 ± 0.00 abc | 1.00 ± 0.00 abc | 4.14 ± 0.46 c | 0.56 ± 0.03 b |
| 0.4 | - | - | 1.43 ± 0.48 abc | 3.29 ± 0.89 ab | 20.71 ± 3.66 ad | 1.24 ± 0.40 b |
| 0.8 | - | - | 1.00 ± 0.22 abc | 1.71 ± 0.47 abc | 5.86 ± 2.19 cd | 0.83 ± 0.17 b |
| 1.6 | - | - | 1.14 ± 0.14 abc | 1.29 ± 0.18 abc | 7.14 ± 2.44 bcd | 0.71 ± 0.19 b |
| 3.2 | - | - | 0.29 ± 0.18 c | 0.43 ± 0.30 b | 1.86 ± 1.70 c | 0.20 ± 0.13 b |
| - | 0.4 | - | 1.86 ± 0.46 ab | 5.86 ± 0.91 a | 26.00 ± 4.34 a | 2.83 ± 0.39 a |
| - | 0.8 | - | 0.86 ± 0.14 abc | 1.71 ± 0.52 abc | 8.00 ± 2.17 bcd | 0.93 ± 0.25 b |
| - | 1.6 | - | 0.14 ± 0.14 c | 0.14 ± 0.14 b | 0.43 ± 0.43 c | 0.07 ± 0.07 b |
| - | 3.2 | - | 0.86 ± 0.14 abc | 1.00 ± 0.22 abc | 8.14 ± 1.97 bcd | 0.64 ± 0.13 b |
| 0.2 | 0.2 | - | 2.00 ± 0.44 a | 3.86 ± 1.12 a | 22.14 ± 4.49 ab | 1.23 ± 0.20 b |
| 0.4 | 0.4 | - | 1.29 ± 0.18 abc | 2.14 ± 0.55 abc | 12.71 ± 4.20 a | 1.11 ± 0.24 b |
| 0.8 | 0.8 | - | 1.43 ± 0.30 abc | 2.14 ± 0.46 abc | 12.43 ± 3.61 a | 1.14 ± 0.31 b |
| 1.6 | 1.6 | - | 1.00 ± 0.22 abc | 1.71 ± 0.61 abc | 8.29 ± 2.73 bcd | 0.91 ± 0.26 b |
| 0.2 | 0.2 | 0.2 | 0.57 ± 0.37 c | 1.00 ± 0.66 abc | 6.00 ± 3.93 cd | 0.51 ± 0.34 b |
| 0.4 | 0.4 | 0.4 | 0.43 ± 0.20 c | 1.29 ± 0.97 abc | 4.43 ± 2.41 c | 0.63 ± 0.38 b |
| Mother Plant | 1st_1 | 1st_2 | 2nd_1 | 2nd_2 | 3rd_1 | 3rd_2 | |
|---|---|---|---|---|---|---|---|
| Mother Plant | |||||||
| 1st_1 | 0.5806 | ||||||
| 1st_2 | 0.5593 | 0.5738 | |||||
| 2nd_1 | 0.4364 | 0.4821 | 0.5714 | ||||
| 2nd_2 | 0.614 | 0.6271 | 0.5 | 0.54 | |||
| 3rd_1 | 0.5625 | 0.6508 | 0.5556 | 0.4912 | 0.6333 | ||
| 3rd_2 | 0.5246 | 0.5645 | 0.5424 | 0.4717 | 0.569 | 0.7368 |
| Contents and Antioxidant Capacity | Obtained Results |
|---|---|
| Total phenolic content | 11.53 GAE g−1 DW |
| Total flavonoid content | 12.99 CE g−1 DW |
| Ferulic acid content | 45.52 mg g−1 DW |
| IC50 with DPPH analysis | 0.43 mg mL−1 |
| IC50 with ABTS+ analysis | 1.99 mg mL−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamdouh, D.; Mahgoub, H.A.M.; Gabr, A.M.M.; Ewais, E.A.; Smetanska, I. Genetic Stability, Phenolic, Flavonoid, Ferulic Acid Contents, and Antioxidant Activity of Micropropagated Lycium schweinfurthii Plants. Plants 2021, 10, 2089. https://doi.org/10.3390/plants10102089
Mamdouh D, Mahgoub HAM, Gabr AMM, Ewais EA, Smetanska I. Genetic Stability, Phenolic, Flavonoid, Ferulic Acid Contents, and Antioxidant Activity of Micropropagated Lycium schweinfurthii Plants. Plants. 2021; 10(10):2089. https://doi.org/10.3390/plants10102089
Chicago/Turabian StyleMamdouh, Diaa, Hany A. M. Mahgoub, Ahmed M. M. Gabr, Emad A. Ewais, and Iryna Smetanska. 2021. "Genetic Stability, Phenolic, Flavonoid, Ferulic Acid Contents, and Antioxidant Activity of Micropropagated Lycium schweinfurthii Plants" Plants 10, no. 10: 2089. https://doi.org/10.3390/plants10102089
APA StyleMamdouh, D., Mahgoub, H. A. M., Gabr, A. M. M., Ewais, E. A., & Smetanska, I. (2021). Genetic Stability, Phenolic, Flavonoid, Ferulic Acid Contents, and Antioxidant Activity of Micropropagated Lycium schweinfurthii Plants. Plants, 10(10), 2089. https://doi.org/10.3390/plants10102089








