Differential Tolerance to Calonectria pseudonaviculata of English Boxwood Plants Associated with the Complexity of Culturable Fungal and Bacterial Endophyte Communities
Abstract
:1. Introduction
2. Results
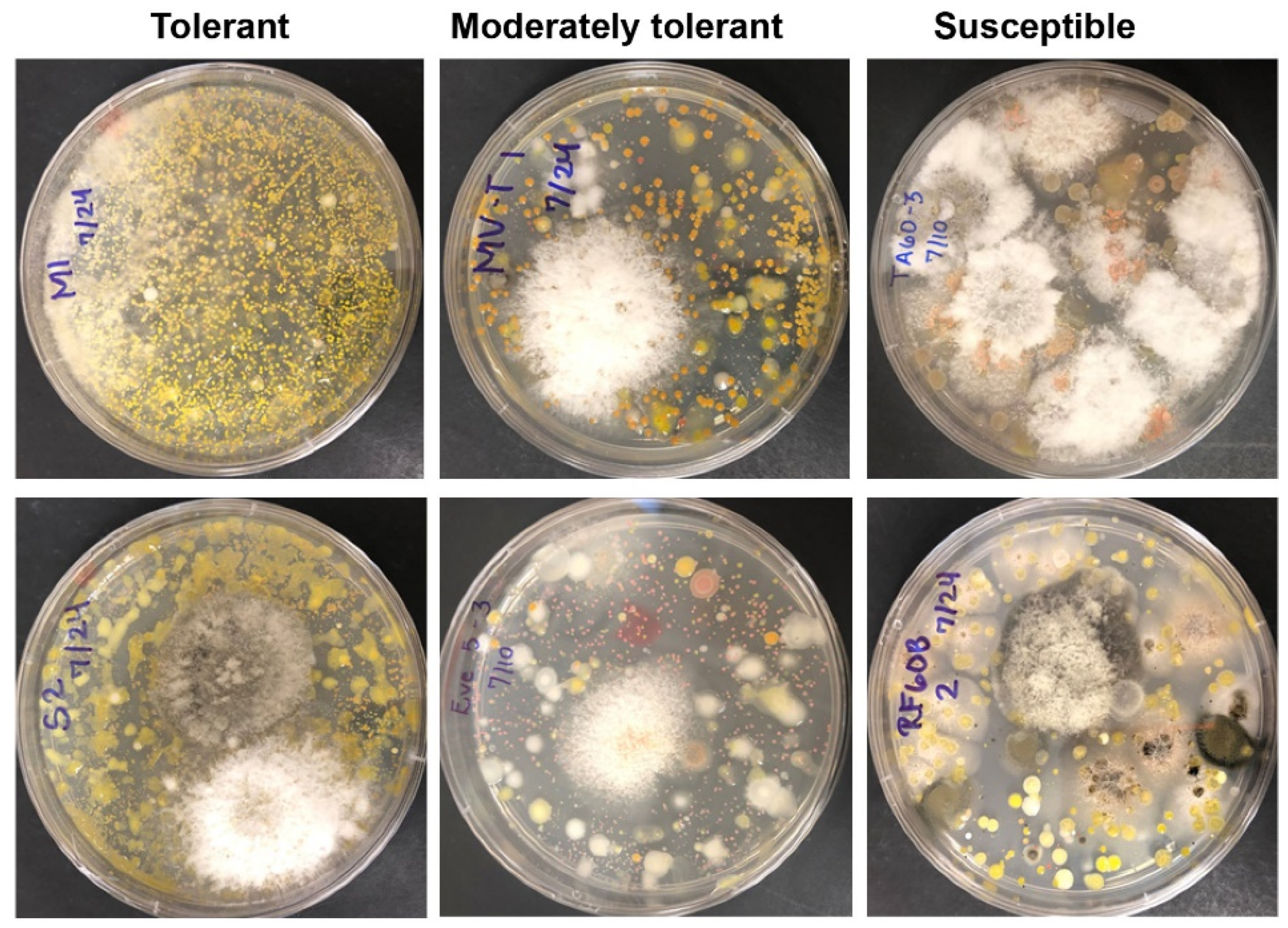
2.1. Blight Tolerance Varied among English Boxwood Plants Sampled
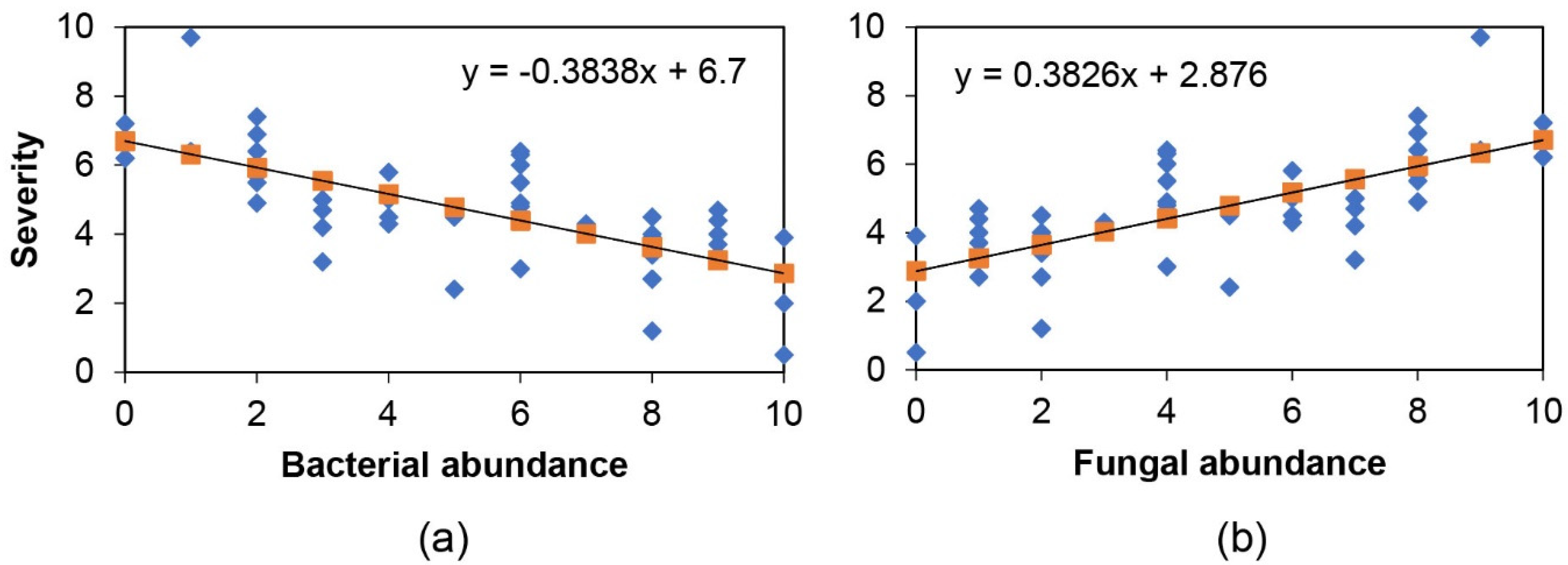
2.2. Endophyte Culture Complexity and Its Blight Tolerance Relation
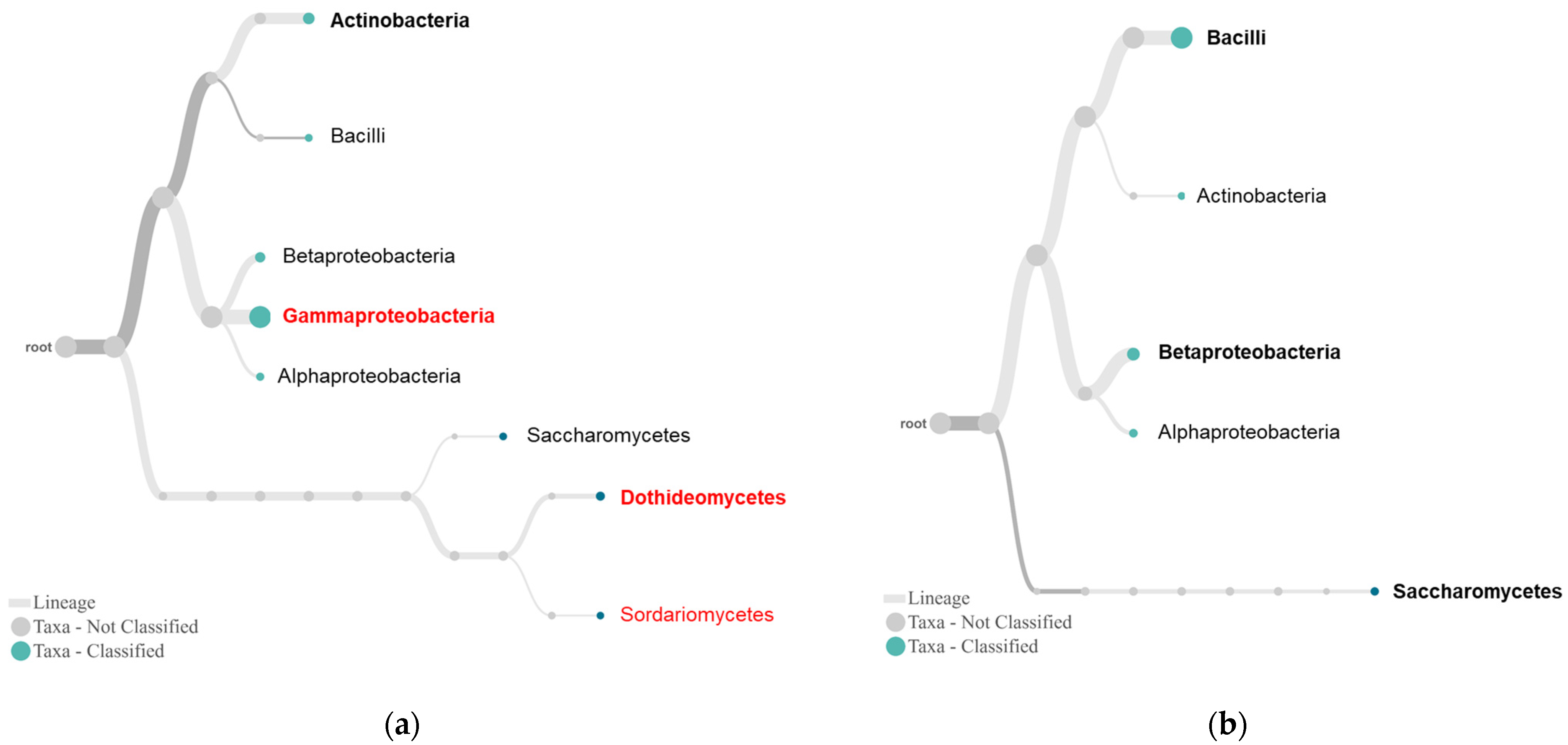
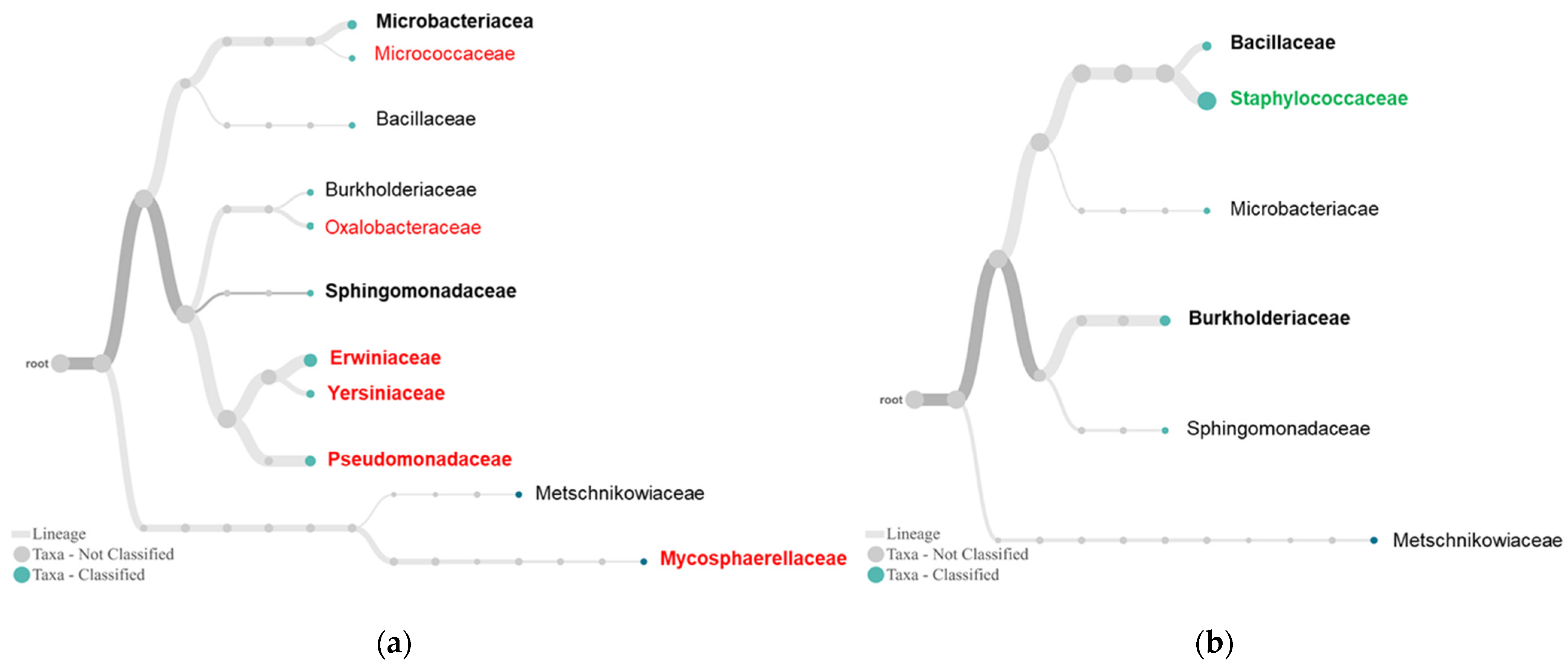
2.3. Differential Complexity of Endophytic Bacterial and Fungal Communities between Tolerant and Susceptible Plants
3. Discussion
4. Materials and Methods
4.1. Plant Samples
4.2. Evaluation of Boxwood Tolerance to Cps
4.3. Endophyte Culture Preparation from Samples
4.4. Evaluation of Cultured Endophyte Complexity
4.5. Endophyte Culture DNA Extraction and Nanopore Sequencing and Analyses
4.6. Data Analysis and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daughtrey, M.L. Boxwood blight: Threat to ornamentals. Annu. Rev. Phytopathol. 2019, 57, 189–209. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, N.; Salgado-Salazar, C.; Crouch, J.A. Boxwood blight: An ongoing threat to ornamental and native boxwood. Appl. Microbiol. Biotechnol. 2018, 102, 4371–4380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C. Fighting plant pathogens together. Science 2019, 365, 229. [Google Scholar] [CrossRef]
- Likins, T.M.; Kong, P.; Avenot, H.F.; Marine, S.; Baudoin, A.; Hong, C. Preventing soil inoculum of Calonectria pseudonaviculata from splashing onto healthy boxwood foliage by mulching. Plant Dis. 2018, 103, 357–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Hong, C.X. Evaluation of biofungicides for control of boxwood blight on boxwood. Plant Dis. Manag. Rep. 2017, 11, OT023. [Google Scholar]
- Kong, P.; Hong, C. A potent Burkholderia endophyte against boxwood blight caused by Calonectria pseudonaviculata. Microorganisms 2020, 8, 310. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Hong, C. Biological control of boxwood blight by Pseudomonas protegens recovered from recycling irrigation systems. Biol. Control 2018, 124, 68–73. [Google Scholar] [CrossRef]
- Kong, P. Evaluation of a novel endophytic Pseudomonas lactis strain for control of boxwood blight. J. Environ. Hortic. 2019, 37, 39–43. [Google Scholar] [CrossRef]
- Kong, P.; Hong, C. Biocontrol of boxwood blight by Trichoderma koningiopsis Mb2. Crop Protect. 2017, 98, 124–127. [Google Scholar] [CrossRef]
- Leggett, M.; Leland, J.; Kellar, K.; Epp, B. Formulation of microbial biocontrol agents—An industrial perspective. Can. J. Plant Pathol. 2011, 33, 101–107. [Google Scholar] [CrossRef]
- Anonymous. Commercial Challenges for Bringing Biopesticides to Market: What Does It Take to Get There? Available online: https://marronebioinnovations.com/commercial-challenges-for-bringing-biopesticides-to-market-what-does-it-take-to-get-there/ (accessed on 13 November 2018).
- Oukala, N.; Aissat, K.; Pastor, V. Bacterial endophytes: The hidden actor in plant immune responses against biotic stress. Plants 2021, 10, 1012. [Google Scholar] [CrossRef] [PubMed]
- Rai, N.; Kumari Keshri, P.; Verma, A.; Kamble, S.C.; Mishra, P.; Barik, S.; Kumar Singh, S.; Gautam, V. Plant associated fungal endophytes as a source of natural bioactive compounds. Mycology 2021, 12, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; Bahar, O.; Papadopoulou, K.K.; Hopkins, D.L.; Obradović, A. Editorial: Role of endophytes in plant health and defense against pathogens. Front. Plant Sci. 2020, 11, 1312. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M. Mechanisms of natural soil suppressiveness to soilborne diseases. Antonie Van Leeuwenhoek 2002, 81, 557–564. [Google Scholar] [CrossRef]
- Gómez Expósito, R.; de Bruijn, I.; Postma, J.; Raaijmakers, J.M. Current Insights into the Role of Rhizosphere Bacteria in Disease Suppressive Soils. Front. Microbiol. 2017, 8, 2529. [Google Scholar] [CrossRef] [PubMed]
- Dini-Andreote, F. Endophytes: The second layer of plant defense. Trends Plant Sci. 2020, 25, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Richardson, P.; Hong, C. Burkholderia sp. SSG is a broad-spectrum antagonist against plant diseases caused by diverse pathogens. Biol. Control 2020, 151, 104380. [Google Scholar] [CrossRef]
- Stewart, J.E.; Kim, M.-S.; Lalande, B.; Klopfenstein, N.B. Chapter 15—Pathobiome and microbial communities associated with forest tree root diseases. In Forest Microbiology; Asiegbu, F.O., Kovalchuk, A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 277–292. [Google Scholar]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; Van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Batdorf, L.R. Boxwood Handbook: A Practical Guide to Knowing and Growing Boxwood, 3rd ed.; Greater Valley Publications, Inc.: Winchester, VA, USA, 2005. [Google Scholar]
- Ganci, M.L.; Benson, D.M.; Ivors, K.L. Susceptibility of commercial boxwood cultivars to Cylindrocladium buxicola, the causal agent of box blight (ABstr.). Phytopathology 2013, 103, 47. [Google Scholar]
- LaMondia, J.A.; Shishkoff, N. Susceptibility of boxwood accessions from the national boxwood collection to boxwood blight and potential for differences between Calonectria pseudonaviculata and C. Henricotiae. HortScience 2017, 52, 873–879. [Google Scholar] [CrossRef]
- Shishkoff, N.; Daughtrey, M.L.; Aker, S.; Olson, R.T. Evaluating boxwood susceptibility to Calonectria pseudonaviculata using cuttings from the national boxwood collection. Plant Health Prog. 2015, 16, 11–15. [Google Scholar] [CrossRef]
- Ryan, C.; Williams-Woodward, J.; Zhang, D. Susceptibility of Sarcococca taxa to boxwood blight caused by Calonectria pseudonaviculata. In Proceedings of the SNA Research Conference 2018, Baltimore, MD, USA, 8–9 January 2018; pp. 62–67. [Google Scholar]
- Rillig, M.C.; Antonovics, J.; Caruso, T.; Lehmann, A.; Powell, J.R.; Veresoglou, S.D.; Verbruggen, E. Interchange of entire communities: Microbial community coalescence. Trends Ecol. Evol. 2015, 30, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Joubert, P.M.; Doty, S.L. Endophytic yeast: Biology, ecology and applications. For. Sci. 2018, 86, 3–14. [Google Scholar] [CrossRef]
- Kong, P.; Hong, C. Complete genome sequence of a boxwood endophyte Burkholderia sp. SSG with broad biotechnological application potential. Biotechnol. Rep. 2020, 26, e00455. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Hong, C. Endophytic Burkholderia sp. SSG as a potential biofertilizer promoting boxwood growth. PeerJ 2020, 8, e9547. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Jourdan, E.; Henry, G.; Duby, F.; Dommes, J.; Barthelemy, J.P.; Thonart, P.; Ongena, M. Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol. Plant-Microbe Interact. 2009, 22, 456–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Community | Abundance | Colony Type | ||
|---|---|---|---|---|
| p-Value | R2 | p-Value | R2 | |
| Bacteria | <0.0001 | 0.4752 | <0.0001 | 0.3755 |
| Fungi | <0.0001 | 0.4796 | 0.2806 | 0.0270 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, P.; Sharifi, M.; Bordas, A.; Hong, C. Differential Tolerance to Calonectria pseudonaviculata of English Boxwood Plants Associated with the Complexity of Culturable Fungal and Bacterial Endophyte Communities. Plants 2021, 10, 2244. https://doi.org/10.3390/plants10112244
Kong P, Sharifi M, Bordas A, Hong C. Differential Tolerance to Calonectria pseudonaviculata of English Boxwood Plants Associated with the Complexity of Culturable Fungal and Bacterial Endophyte Communities. Plants. 2021; 10(11):2244. https://doi.org/10.3390/plants10112244
Chicago/Turabian StyleKong, Ping, Melissa Sharifi, Adria Bordas, and Chuanxue Hong. 2021. "Differential Tolerance to Calonectria pseudonaviculata of English Boxwood Plants Associated with the Complexity of Culturable Fungal and Bacterial Endophyte Communities" Plants 10, no. 11: 2244. https://doi.org/10.3390/plants10112244
APA StyleKong, P., Sharifi, M., Bordas, A., & Hong, C. (2021). Differential Tolerance to Calonectria pseudonaviculata of English Boxwood Plants Associated with the Complexity of Culturable Fungal and Bacterial Endophyte Communities. Plants, 10(11), 2244. https://doi.org/10.3390/plants10112244






