Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.)
Abstract
:1. Introduction
2. Results
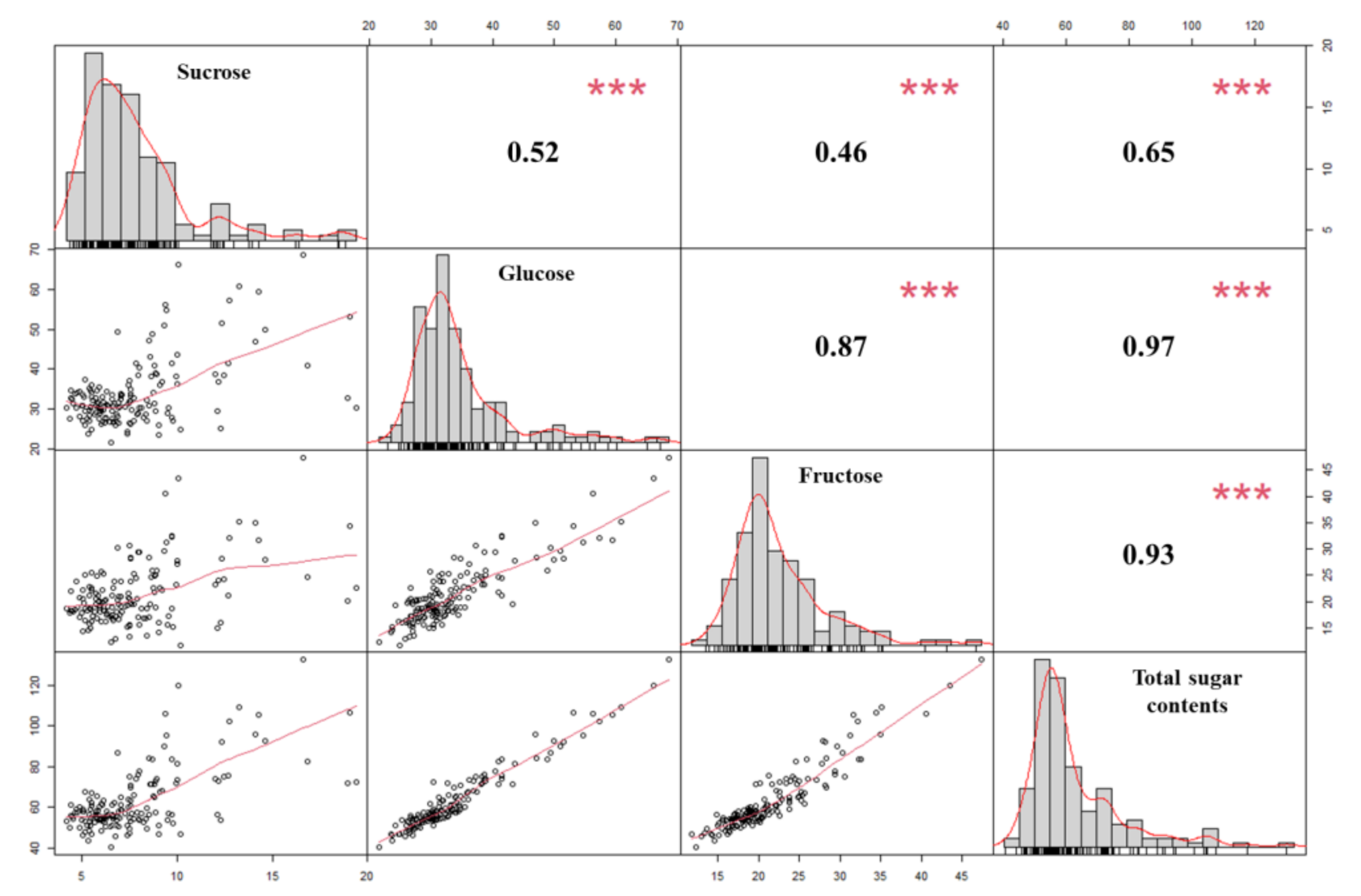
2.1. Sugar Content Analysis of Onion Samples
2.2. SNP Detection and Genotyping-by-Sequencing (GBS) Analysis
2.3. Construction of an Onion Genetic Linkage Map
2.4. Comparison of the ‘16P118 × Sweet Green’ and ‘SP3B × H6’ Onion Genetic Linkage Maps
2.5. Identification of QTLs for Sugar Content in Onion
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Assessment of Sugar Content
4.3. DNA Extraction
4.4. Genotyping-by-Sequencing Analysis
4.5. Genetic Linkage Construction
4.6. QTL Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BLASTN | Basic Local Alignment Search Tool Nucleotide |
| CAPS | cleaved amplified polymorphic sequence |
| CIM | composite interval mapping |
| cM | centi Morgan |
| FC | fructose content |
| GBS | genotyping-by-sequencing |
| GC | glucose content |
| LOD | logarithm of Odds |
| MAS | marker-assisted selection |
| NGS | next-generation sequencing |
| QTL | quantitative trait loci |
| RAPD | random amplified polymorphic DNA |
| RE | restriction enzymes |
| RFLP | restriction fragment length polymorphism |
| SC | sucrose content |
| SNP | single nucleotide polymorphism |
| SSR | simple sequence repeats |
| TSC | total sugar content |
References
- Khosa, J.S.; McCallum, J.; Dhatt, A.S.; Macknght, R.C. Enhancing onion breeding using molecular tools. Plant Breed. 2016, 135, 9–20. [Google Scholar] [CrossRef]
- Jo, J.; Purushotham, P.M.; Han, K.; Lee, H.R.; Nah, G.; Kang, B.C. Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays. Front. Plant Sci. 2017, 8, 1606. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/ (accessed on 10 October 2018).
- Sims, C.A.; Golaszewski, R. Vegetable flavor and changes during postharvest storage. In Postharvest Physiology and Pathology of Vegetable; Bartz, J.A., Brecht, J.K., Eds.; Marcel Dekker: New York, NY, USA, 2003; pp. 331–340. [Google Scholar]
- Joesten, M.D.; Hogg, J.L.; Castellion, M.E. Sweetness relative to sucrose. In The World of Chemistry: Essentials, 4th ed.; Thomson Books: Belmont, CA, USA, 2007; p. 359. [Google Scholar]
- VandenLangenberg, K.M.; Bethke, P.C.; Nienhuis, J. Patterns of fructose, glucose, and sucrose accumulation in snap and dry bean (Phaseolus vulgaris) pods. HortScience 2012, 47, 874–878. [Google Scholar] [CrossRef]
- Darbyshire, B.; Henry, R.J. The distribution of fructans in onions. New Phytol. 1978, 81, 29–34. [Google Scholar] [CrossRef]
- Vavrina, C.S.; Smittle, D.A. Evaluating sweet onion cultivars for sugar concentrations and pungency. HortScience 1993, 28, 804–806. [Google Scholar] [CrossRef]
- Ritsema, T.; Smeekens, S. Fructans: Beneficial for plants and humans. Curr. Opin. Plant Biol. 2003, 6, 223–230. [Google Scholar] [CrossRef]
- McCallum, J.; Clarke, A.; Pither-Joyce, M.; Shaw, M.; Butler, R.; Brash, R.; Scheffer, J.; Sims, I.; van Heusden, S.; Shigyo, M.; et al. Genetic mapping of a major gene affecting onion bulb fructan content. Theor. Appl. Genet. 2006, 112, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Finkers, R.; Kaauwen, M.; Ament, K.; Burger-Meijer, K.; Egging, R.; Huits, H.; Kodde, L.; Kroon, L.; Shigyo, M.; Sato, S.; et al. Insights form the first genome assembly of Onion (Allium cepa). bioRxiv 2021. [Google Scholar] [CrossRef]
- King, J.J.; Bradeen, J.M.; Bark, O.; McCallum, J.A.; Havey, M.J. A low-density genetic map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome. Theor. Appl. Genet. 1998, 96, 52–62. [Google Scholar] [CrossRef]
- Van Heusden, A.W.; Van Ooijen, J.W.; Ginkel, R.V.-V.; Verbeek, W.H.J.; Wietsma, W.A.; Kik, C. A genetic map of an interspecific cross in Allium based on amplified fragment length polymorphism (AFLPTM) markers. Theor. Appl. Genet. 2000, 100, 118–126. [Google Scholar] [CrossRef]
- Van Heusden, A.W.; Shigyo, M.; Tashiro, Y.; Ginkel, R.V.-V.; Kik, C. AFLP linkage group assignment to the chromosomes of Allium cepa L. via monosomic addition lines. Theor. Appl. Genet. 2000, 100, 480–486. [Google Scholar] [CrossRef]
- McCallum, J.; Leite, D.; Pither-Joyce, M.; Havey, M.J. Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L.). Theor. Appl. Genet. 2001, 103, 979–991. [Google Scholar] [CrossRef]
- Jakse, J.; Martin, W.; McCallum, J.; Havey, M.J. Single nucleotide polymorphisms, Indels, and simple sequence repeats for onion (Allium cepa L.) cultivar identification. J. Am. Soc. Hortic. Sci. 2005, 130, 912–917. [Google Scholar] [CrossRef]
- Baldwin, S.; Pither-Joyce, M.; Wright, K.; Chen, L.; McCallum, J. Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Mol. Breed. 2012, 30, 1401–1411. [Google Scholar] [CrossRef]
- Hayward, A.; Mason, A.S.; Dalton-Morgan, J.; Zander, M.; Edwards, D.; Batley, J. SNP discovery and applications in Brassica napus. J. Plant Biotechnol. 2012, 39, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [Green Version]
- Pootakham, W.; Ruang-Aeerate, P.; Jomchai, N.; Sonthirod, C.; Sangsrakru, D.; Yoocha, T.; Theerawattanasuk, K.; Nirapathpongporn, K.; Romruensukharom, P.; Tragoonrung, S.; et al. Construction of a high-density integrated genetic linkage map of rubber tree (Hevea brasiliensis) using genotyping-by-sequencing (GBS). Front. Plant Sci. 2015, 6, 367. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.S.; Choi, S.C.; Jun, T.H.; Kim, C. Genotyping-by-sequencing: A Promising tool for plant genetics research and breeding. Hortic. Environ. Biotechnol. 2017, 58, 425–431. [Google Scholar] [CrossRef]
- Han, K.; Lee, H.Y.; Ro, N.Y.; Hur, O.S.; Lee, J.H.; Kwon, J.K.; Kang, B.C. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 2018, 16, 1546–1558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddique, M.I.; Lee, H.Y.; Ro, N.Y.; Han, K.; Venkatesh, J.; Solomon, A.M.; Patil, A.S.; Changkwian, A.; Kwon, J.K.; Kang, B.C. Identifying candidate genes for Phytophthora capsici resistance in pepper (Capsicum annuum) via genotyping-by-sequencing-based QTL mapping and genome-wide association study. Sci. Rep. 2019, 9, 9962. [Google Scholar] [CrossRef]
- Solomon, A.M.; Han, K.; Lee, J.H.; Lee, H.Y.; Jang, S.; Kang, B.C. Genetic diversity and population structure of Ethiopian Capsicum germplasms. PLoS ONE 2019, 14, e0216886. [Google Scholar] [CrossRef]
- He, J.; Zhan, X.; Laroche, A.; Lu, Z.X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Kim, S.; Lee, J. Construction of an onion (Allium cepa L.) genetic linkage map using genotyping-by-sequencing analysis with a reference gene set and identification of QTLs controlling anthocyanin synthesis and content. Plants 2020, 9, 616. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, M.S.; Kim, Y.M.; Yeom, S.I.; Cheong, K.; Kim, K.T.; Jeon, J.; Kim, S.; Kim, D.S.; Sohn, S.H.; et al. Integrative structural annotation of denovo RNA-Seq provides an accurate reference gene set of the enormous genome of the onion (Allium cepa L.). DNA Res. 2015, 22, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Labate, J.A.; Glaubitz, J.C.; Havey, M.J. Genotyping by sequencing for SNP marker development in onion. Genome 2020, 63, 607–613. [Google Scholar] [CrossRef] [PubMed]
- McCallum, J.; Baldwin, S.; Macknight, R.; Khosa, J.; Shaw, M. Molecular mapping of genes and QTL: Progress to date and development of new population resources for NGS genetics. In The Allium Genomes; Sshigyo, M., Khar, A., Abdelrahman, M., Eds.; Compendium of Plant Genomes; Springer: Cham, Switzerland, 2018; pp. 181–196. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Comertpay, G.; Yildiz, R.H.; Ahmad, F.; Alsaleh, A.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, S.; Revanna, R.; Thomson, S.; Pither-Joyce, M.; Wright, K.; Crowhurst, R.; Fiers, M.; Chen, L.; Macknight, R.; McCallum, J. A toolkit for bulk PCR-based marker design from next-generation sequence data: Application for development of a framework linkage map in bulb onion (Allium cepa L.). BMC Genom. 2012, 13, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duangjit, J.; Bohanec, B.; Chan, A.P.; Town, C.D.; Havey, M.J. Transcriptome sequencing to produce SNP-based genetic maps of onion. Theor. Appl. Genet. 2013, 126, 2093–2101. [Google Scholar] [CrossRef]
- Lee, E.J.; Yoo, K.S.; Jifon, J.; Patil, B.S. Characterization of shortday onion cultivars of 3 pungency levels with flavor precursor, free amino acid, sulfur, and sugar contents. J. Food Sci. 2009, 74, C475–C480. [Google Scholar] [CrossRef]
- Darbyshire, B. Changes in the carbohydrate content of onion bulbs stored for various times at different temperatures. J. Hortic. Sci. 1978, 53, 195–201. [Google Scholar] [CrossRef]
- Havey, M.J.; Galmarini, C.R.; Gökçe, A.F.; Henson, C. QTL affecting soluble carbohydrate concentrations in stored onion bulbs and their association with flavor and health-enhancing attributes. Genome 2004, 47, 463–468. [Google Scholar] [CrossRef]
- Raines, S.; Henson, C.; Havey, M.J. Genetic analyses of soluble carbohydrate concentrations in onion bulbs. J. Am. Soc. Hortic. Sci. 2009, 134, 618–623. [Google Scholar] [CrossRef]
- McCallum, J.; Pither-Joyce, M.; Shaw, M.; Kenel, F.; Davis, S.; Butler, R.; Scheffer, J.; Jakse, J.; Havey, M.J. Genetic mapping of sulfur assimilation genes reveals a QTL for onion bulb pungency. Theor. Appl. Genet. 2007, 114, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Ko, E.Y.; Assefa, A.D.; Ha, S.; Nile, S.H.; Lee, E.T.; Park, S.W. Temperature-dependent studies on the total phenolics, flavonoids, antioxidant activities, and sugar content in six onion varieties. J. Food Drug Anal. 2015, 23, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, F.; Terry, L.A.; Chope, G.A.; Faul, C.F. Effect of extraction procedure on measured sugar concentrations in onion (Allium cepa L.) bulbs. J. Agric. Food Chem. 2007, 55, 4299–4306. [Google Scholar] [CrossRef]
- Galdón, B.R.; Rodríguez, C.T.; Rodríguez, E.R.; Romero, C.D. Fructans and major compounds in onion cultivars (Allium cepa). J. Food Compos. Anal. 2009, 22, 25–32. [Google Scholar] [CrossRef]
- Benkeblia, N.; Varoquaux, P. Effects of gamma irradiation, temperature and storage time on the status of the glucose, fructose and sucrose in onion bulbs Allium cepa L. Int. Agrophys. 2003, 17, 1–5. [Google Scholar]
- Choi, M.H.; Choi, H.J.; Hong, S.S.; Lim, B.S. Effects of gradual cooling treatment on the skin blackening and physicochemical characteristics of ‘Chuhwang’ pear fruit. Korean J. Food Preserv. 2020, 27, 145–158. [Google Scholar] [CrossRef]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Manivannan, A.; Choi, S.; Jun, T.H.; Yang, E.Y.; Kim, J.H.; Lee, E.S.; Lee, H.E.; Kim, D.S.; Ahn, Y.K. Genotyping by Sequencing-Based Discovery of SNP Markers and Construction of Linkage Map from F5 Population of Pepper with Contrasting Powdery Mildew Resistance Trait. BioMed Res. Int. 2021, 2021, 1–8. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.E.; Oh, S.K.; Lee, B.M.; Jo, S.H. Genome-wide SNP calling using next generation sequencing data in tomato. Mol. Cells 2014, 37, 36–42. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distance from recombination value. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, North CarolinaState University: Raleigh, NC, USA, 2007. [Google Scholar]
- Peterson, B.G.; Carl, P.; Boudt, K.; Bennett, R.; Ulrich, J.; Zivot, E.; Cornily, D.; Hung, E.; Lestel, M.; Balissoon, K.; et al. Package ‘Performance Analytics’; R Team Cooperation: Vienna, Austria, 2020. [Google Scholar]




| Sucrose | Glucose | Fructose | Total Sugar | ||
|---|---|---|---|---|---|
| 16P118 (female) | 7.6 ± 0.4 a 1 | 31.9 ± 1.3 a | 23.8 ± 1.3 a | 63.2 ± 2.2 a | |
| Sweet Green (male) | 19.4 ± 3.3 b | 30.4 ± 5.4 a | 22.5 ± 3.7 a | 72.3 ± 4.1 a | |
| Offspring | Range | 3.7 to 22.3 | 8.6 to 73.3 | 9.1 to 51.3 | 26.5 to 135.2 |
| Average | 7.6 | 33.4 | 21.0 | 62.1 | |
| Summary of Illumina Sequencing | Data |
|---|---|
| Number of plants for multiplexing | 156 |
| Total number of raw reads generated | 1,373,106,298 |
| Total base number of raw reads (bp) | 207,339,050,998 (207.3 Gbp) |
| Total number of demultiplexed reads | 1,249,719,142 (91.0%) |
| Total number of trimmed reads | 1,170,326,476 (85.2%) |
| Total number of mapped reads | 565,805,698(41.2%) |
| Total number of mapped regions | 1,638,943 |
| Average depth of mapped region | 147.77 |
| Total length of mapped regions (bp) | 1,680,157 (1.7 Mbp) |
| Total length of the reference gene set (bp) | 280,586,290 (280.6 Mbp) |
| Coverage of the reference gene set | 0.7676% |
| Total number of SNPs detected | 24,341 |
| Total number of SNPs filtered | 815 |
| Chromosome No. | Number of SNPs | Length of Linkage Distance (cM) |
|---|---|---|
| 1 | 33 | 133.8 |
| 2 | 39 | 135.8 |
| 3 | 37 | 152.7 |
| 4 | 17 | 88.8 |
| 5 | 26 | 119.0 |
| 6 | 20 | 50.8 |
| 7 | 26 | 55.3 |
| 8 | 18 | 90.7 |
| Total | 216 | 827.0 |
| Trait | QTL Name | Chr. | Peak Position 1 (cM) | LOD 2 | LOD Threshold 3 | Additive Effect | Dominance Effect | R2 (%) 4 |
|---|---|---|---|---|---|---|---|---|
| Sucrose | qSC4.1 | 4 | 31.8 | 5.37 | 3.48 | 1.15 | 0.78 | 11.47 |
| Glucose | qGC5.1 | 5 | 73.0 | 4.63 | 3.52 | 3.12 | −1.51 | 16.87 |
| Fructose | qFC5.1 | 5 | 70.4 | 4.15 | 3.44 | 1.95 | −1.21 | 15.39 |
| Total sugar | qTSC5.1 | 5 | 69.3 | 3.41 | 3.40 | 4.84 | −2.92 | 6.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-R.; Kim, C.W.; Han, J.; Choi, H.J.; Han, K.; Lee, E.S.; Kim, D.-S.; Lee, J.; Siddique, M.I.; Lee, H.-E. Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.). Plants 2021, 10, 2267. https://doi.org/10.3390/plants10112267
Lee Y-R, Kim CW, Han J, Choi HJ, Han K, Lee ES, Kim D-S, Lee J, Siddique MI, Lee H-E. Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.). Plants. 2021; 10(11):2267. https://doi.org/10.3390/plants10112267
Chicago/Turabian StyleLee, Ye-Rin, Cheol Woo Kim, JiWon Han, Hyun Jin Choi, Koeun Han, Eun Su Lee, Do-Sun Kim, Jundae Lee, Muhammad Irfan Siddique, and Hye-Eun Lee. 2021. "Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.)" Plants 10, no. 11: 2267. https://doi.org/10.3390/plants10112267
APA StyleLee, Y.-R., Kim, C. W., Han, J., Choi, H. J., Han, K., Lee, E. S., Kim, D.-S., Lee, J., Siddique, M. I., & Lee, H.-E. (2021). Genotyping-by-Sequencing Derived Genetic Linkage Map and Quantitative Trait Loci for Sugar Content in Onion (Allium cepa L.). Plants, 10(11), 2267. https://doi.org/10.3390/plants10112267









