The Influence of Hop Latent Viroid (HLVd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus lupulus L.) Glandular Trichomes
Abstract
:1. Introduction
2. Results
2.1. Contents of Secondary Metabolites in Maturated Hop Cones of Healthy and HVLd-Infected Plants
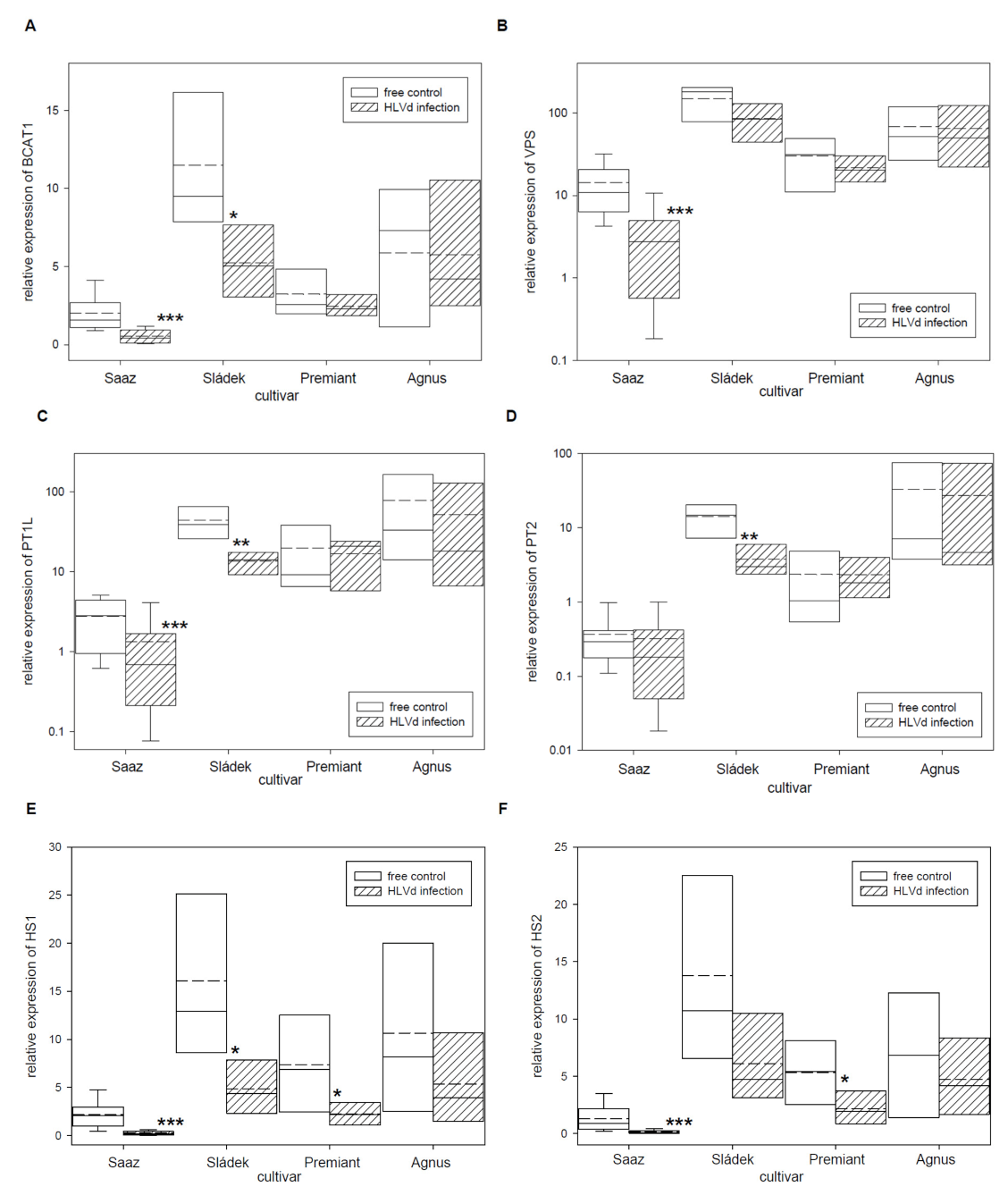
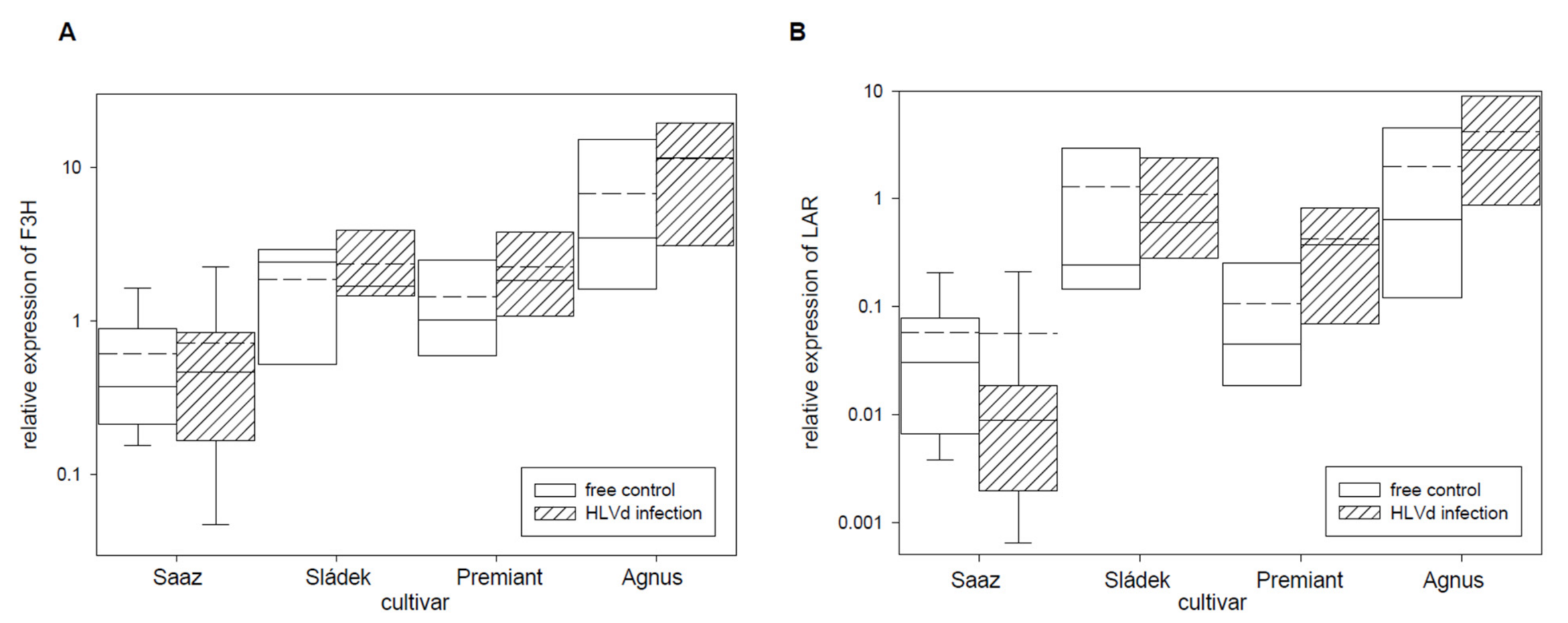
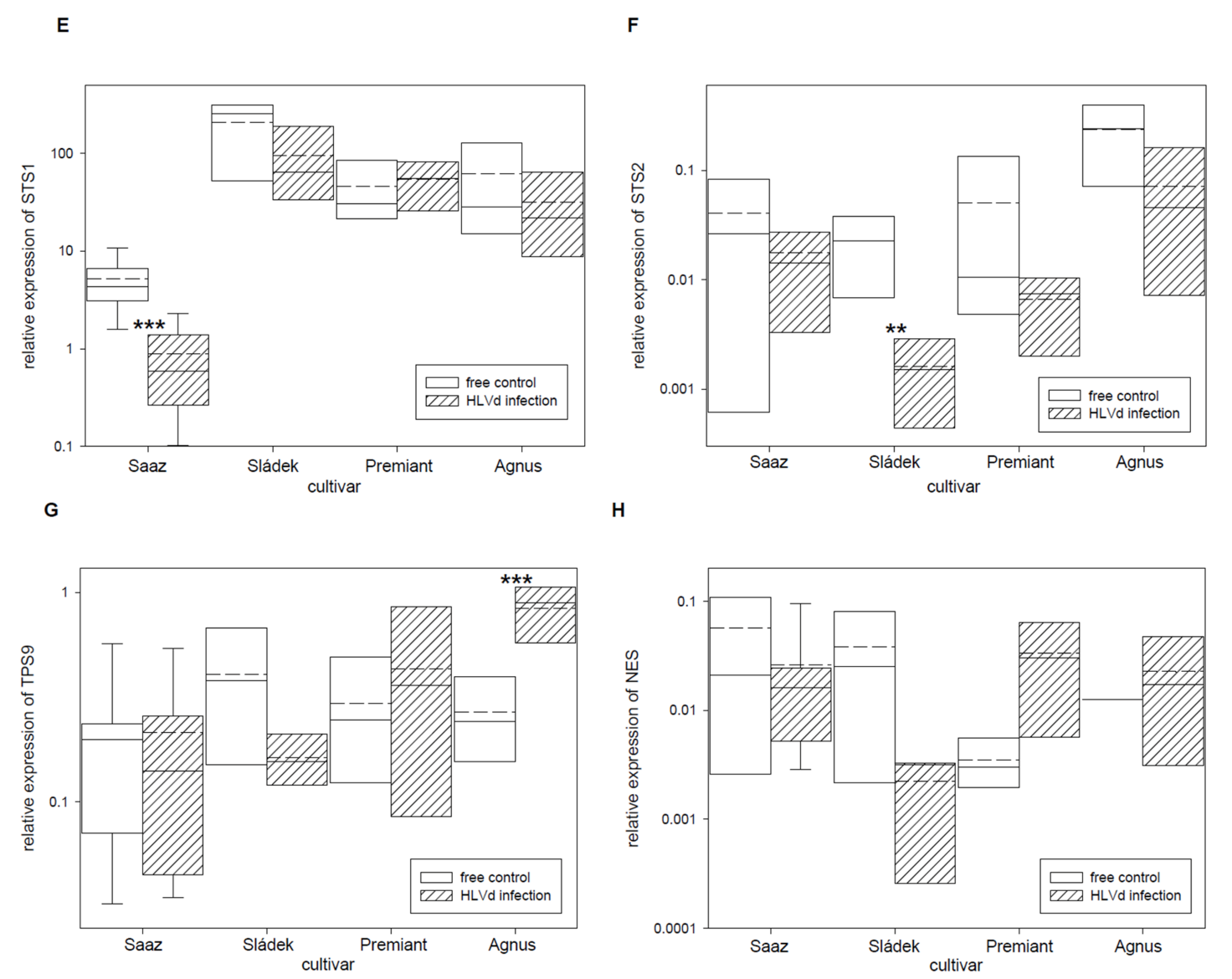
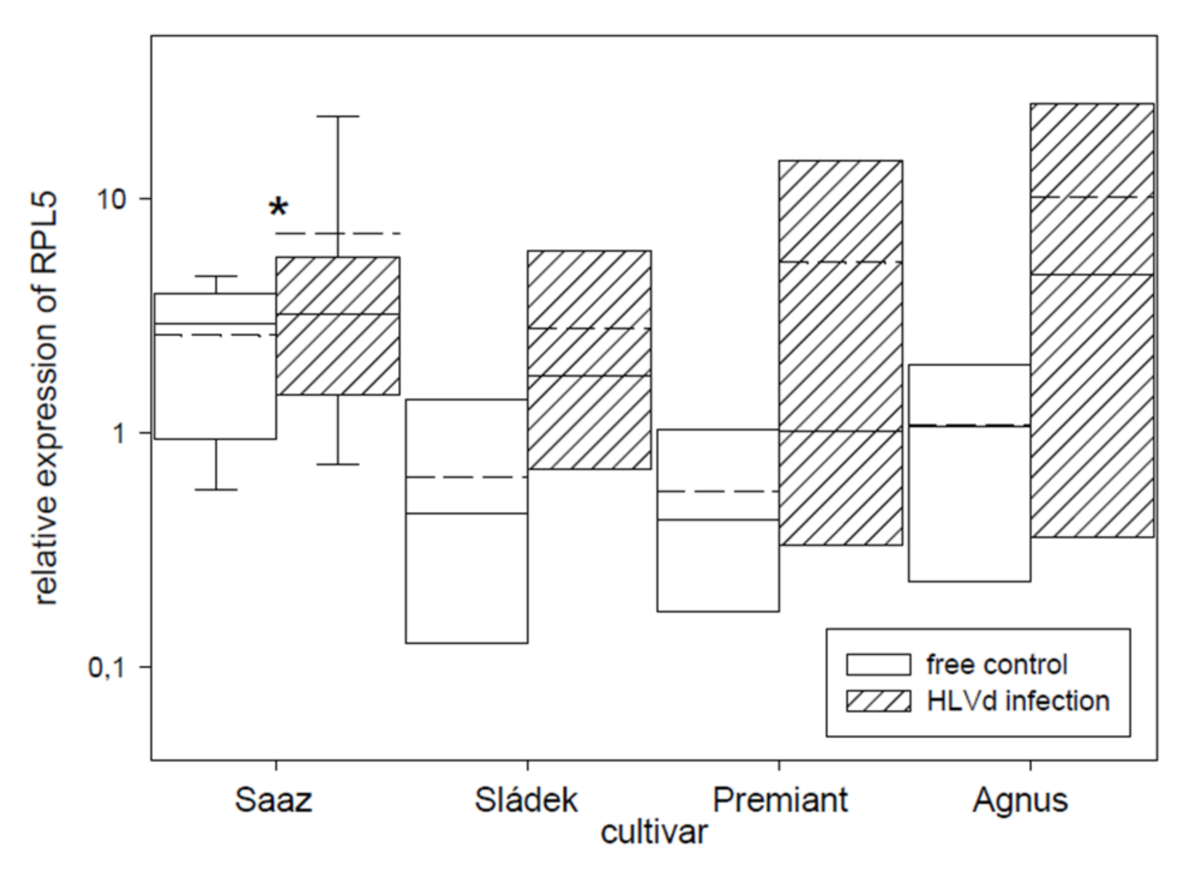
2.2. Analyses of Gene Expressions in Hop Cones of Healthy and HVLd-Infected Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Hop Latent Viroid (HLVd) Detection
4.2. Chemical Analyses
4.3. Gene Expression Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hadidi, A.; Flores, R.; Randles, J.W.; Palukaitis, P. Viroids and Sattelites; Academic Press: Cambridge, UK, 2017; p. 716. ISBN 978-0-12-801498-1. [Google Scholar]
- Pethybridge, S.J.; Hay, F.S.; Barbara, D.J.; Eastwell, K.C.; Wilson, C.R. Viruses and viroid infected hop: Epidemiology and management. Plant Dis. 2008, 92, 324–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sano, T.; Yoshida, H.; Goshono, M.; Monma, T.; Kawasaki, H.; Ishizaki, K. Characterization of a new viroid strain from hops: Evidence for viroid speciation by isolation in different host species. J. Gen. Plant Pathol. 2004, 70, 181–187. [Google Scholar] [CrossRef]
- Radišek, S.; Majer, A.; Jakše, J.; Javornik, B.; Matoušek, J. First report of Hop stunt viroid infecting hop in Slovenia. Plant Dis. 2012, 96, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Kappagantu, M.; Nelson, M.E.; Bullock, J.M.; Kenny, S.T.; Eastwell, K.C. Hop stunt viroid: Effects on vegetative growth and yield of hop cultivars, and its cistribution in Central Washington state. Plant Dis. 2017, 101, 607–612. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.M.; Vaitheeswaran, V.; Ambrose, S.J.; Purves, R.W.; Page, J.E. Transcriptome analysis of bitter acid biosynthesis and precursor pathways in hop (Humulus lupulus). BMC Plant Biol. 2013, 13, 12. [Google Scholar] [CrossRef] [Green Version]
- Radišek, S.; Oset, M.; Čerenak, A.; Jakše, J.; Knapič, V.; Matoušek, J.; Javornik, B. Research activities focused on hop viroid diseases in Slovenia. In Proceedings of the Scientific Commission of IHGC, Kiev, Ukraine, 4–9 June 2013; pp. 58–3785. [Google Scholar]
- Radišek, S. Management of hop viroid diseases in Slovenia. In Proceedings of the 56th International Hop Growers’ Convention (IHGC) Congress, Yakima, WA, USA, 30 July–3 August 2017. [Google Scholar]
- Patzak, J.; Matoušek, J.; Krofta, K.; Svoboda, P. Hop latent viroid (HLVd)-caused pathogenesis: Effects of HLVd infection on lupulin composition of meristem culture-derived hop (Humulus lupulus L.). Biol. Plant. 2001, 44, 579–585. [Google Scholar] [CrossRef]
- Seigner, E.; Haugg, B.; Hager, P.; Enders, R.; Kneidl, J.; Lutz, A.; Seigner, L.; Einberger, K.; Absmeier, C.; Keckel, L.; et al. Realtime PCR based diagnostics and meristem culture—Essential tools for healthy hops. In Proceedings of the Scientific Commission of IHGC, Bischoffsheim, France, 7–11 July 2019; pp. 114–3785. [Google Scholar]
- Barbara, D.J.; Morton, A.; Adams, A.N.; Green, C.P. Some effects of hop latent viroid on two cultivars of hop (Humulus lupulus) in the UK. Ann. Appl. Biol. 1990, 117, 359–366. [Google Scholar] [CrossRef]
- Adams, A.N.; Barbara, D.J.; Morton, A. Effects of hop latent viroid on weight and quality of the cones of the hop cultivar Wye challenger. Ann. Appl. Biol. 1991, 118 (Suppl.), 126–127. [Google Scholar]
- Adams, A.N.; Morton, A.; Barbara, D.; Ridout, M. The distribution and spread of hop latent viroid within two commercial plantings of hop (Humulus lupulus). Ann. Appl. Biol. 1992, 121, 585–592. [Google Scholar] [CrossRef]
- Pistelli, L.; Ferri, B.; Cioni, P.L.; Koziara, M.; Agacka, M.; Skomra, U. Aroma profile and bitter acid characterization of hop cones (Humulus lupulus L.) of five healthy and infected Polish cultivars. Ind. Crops Prod. 2018, 124, 653–662. [Google Scholar] [CrossRef]
- Kovačevič, M.; Kač, M. Determination and verification of hop varieties by analysis of essential oils. Food Chem. 2002, 77, 489–494. [Google Scholar] [CrossRef]
- Okada, Y.; Ito, K. Cloning and analysis of valerophenone synthase gene expressed specifically in lupulin gland of hop (Humulus lupulus L.). Biosci. Biotechnol. Biochem. 2001, 65, 150–155. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, F.; Liu, B.; Huhman, D.V.; Sumner, L.W.; Dixon, R.A.; Wang, G. Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol. Plant 2013, 6, 1301–1317. [Google Scholar] [CrossRef] [PubMed]
- Okada, K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011, 75, 1219–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsurumaru, Y.; Sasaki, K.; Miyawaki, T.; Momma, T.; Umemoto, N.; Yazaki, K. An aromatic prenyltransferase-like gene HlPT-1 preferentially expressed in lupulin glands of hop. Plant Biotechnol. 2010, 27, 199–204. [Google Scholar] [CrossRef] [Green Version]
- Tsurumaru, Y.; Sasaki, K.; Miyawaki, T.; Uto, Y.; Momma, T.; Umemoto, N.; Yazaki, K. HlPT-1, a membrane-bound prenyltransferase responsible for the biosynthesis of bitter acids in hops. Biochem. Biophys. Res. Comm. 2012, 417, 393–398. [Google Scholar] [CrossRef]
- Li, H.; Ban, Z.; Qin, H.; Ma, L.; King, A.J.; Wang, G. A heteromeric membrane-bound prenyltransferase complex from hop catalyzes three sequential aromatic prenylations in the bitter acid pathway. Plant Physiol. 2015, 167, 650–659. [Google Scholar] [CrossRef]
- Champagne, A.; Boutry, M. A comprehensive proteome map of glandular trichomes of hop (Humulus lupulus L) female cones: Identification of biosynthetic pathways of the major terpenoid-related compounds and possible transport proteins. Proteomics 2017, 17, 1600411. [Google Scholar] [CrossRef]
- Okada, Y.; Sugimoto, M.; Ito, K. Molecular cloning and expression of farnesyl pyrophosphate synthase gene responsible for essential oil biosynthesis in hop (Humulus lupulus). J. Plant Physiol. 2001, 158, 1183–1188. [Google Scholar] [CrossRef]
- Wang, G.; Tian, L.; Aziz, N.; Broun, P.; Dai, X.; He, J.; King, A.; Zhao, P.X.; Dixon, R.A. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 2008, 148, 1254–1266. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Dixon, R.A. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Nat. Acad. Sci. USA 2009, 106, 9914–9919. [Google Scholar] [CrossRef] [Green Version]
- Matoušek, J.; Novák, P.; Bříza, J.; Patzak, J.; Niedermaierová, H. Cloning and characterisation of chs-specific DNA and cDNA sequences from hop (Humulus lupulus L.). Plant Sci. 2002, 162, 1007–1018. [Google Scholar] [CrossRef]
- Ban, Z.; Qin, H.; Mitchell, A.J.; Liu, B.; Zhang, F.; Weng, J.-K.; Dixon, R.A.; Wang, G. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc. Nat. Acad. Sci. USA 2018, 115, E5223–E5232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, J.; Culley, L.K.; Lu, Y.; Liu, E.; Matthews, P.D.; Stevens, J.F.; Page, J.E. EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell 2008, 20, 186–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoušek, J.; Kocábek, T.; Patzak, J.; Füssy, Z.; Procházková, J.; Heyerick, A. Combinatorial analysis of lupulin gland transcription factors from R2R3Myb, bHLH and WDR families indicates a complex regulation of chs_H1 genes essential for prenylflavonoid biosynthesis in hop (Humulus lupulus L.). BMC Plant Biol. 2012, 12, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.K.; Duraisamy, G.S.; Khare, M.; Kocábek, T.; Jakse, J.; Bříza, J.; Patzak, J.; Sano, T.; Matoušek, J. Genome-wide transcriptome profiling of transgenic hop (Humulus lupulus L.) constitutively overexpressing HlWRKY1 and HlWDR1 transcription factors. BMC Genom. 2018, 19, 739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoušek, J.; Kocábek, T.; Patzak, J.; Bříza, J.; Siglová, K.; Mishra, A.K.; Duraisamy, G.S.; Týcová, A.; Ono, E.; Krofta, K. The “putative” role of transcription factors from HlWRKY family in the regulation of the final steps of prenylflavonid and bitter acids biosynthesis in hop (Humulus lupulus L.). Plant Mol. Biol. 2016, 92, 263–277. [Google Scholar] [CrossRef]
- Kocábek, T.; Mishra, A.K.; Matoušek, J.; Patzak, J.; Lomnická, A.; Khare, M.; Krofta, K. The R2R3 transcription factor HlMYB8 and its role in flavonoid biosynthesis in hop (Humulus lupulus L.). Plant Sci. 2018, 269, 32–46. [Google Scholar] [CrossRef]
- Pokorn, T.; Radišek, S.; Javornik, B.; Štajner, N.; Jakše, J. Development of hop transcriptome to support research into host-viroid interactions. PLoS ONE 2017, 12, e0184528. [Google Scholar] [CrossRef] [Green Version]
- Štajner, N.; Radišek, S.; Mishra, A.K.; Nath, V.S.; Matoušek, J.; Jakše, J. Evaluation of disease severity and global transcriptome response induced by citrus bark cracking viroid, hop latent viroid, and their co-Infection in hop (Humulus lupulus L.). Int. J. Mol. Sci. 2019, 20, 3154. [Google Scholar] [CrossRef] [Green Version]
- Nath, V.S.; Shrestha, A.; Awasthi, P.; Mishra, A.K.; Kocábek, T.; Matoušek, J.; Sečnik, A.; Jakše, J.; Radišek, S.; Hallan, V. Mapping the gene expression spectrum of mediator subunits in response to viroid infection in plants. Int. J. Mol. Sci. 2020, 21, 2498. [Google Scholar] [CrossRef] [Green Version]
- Eiras, M.; Nohales, M.A.; Kitajima, E.W.; Flores, R.; Daròs, J.A. Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically Potato spindle tuber viroid RNA. Arch. Virol. 2010, 156, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qu, J.; Ji, S.; Wallace, A.J.; Wu, J.; Li, Y.; Gopalan, V.; Ding, B. A land plant-specific transcription factor directly enhances transcription of a pathogenic noncoding RNA template by DNA-dependent RNA Polymerase II. Plant Cell 2016, 28, 1094–1107. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Smith, H.N.; Ren, D.; Mudiyanselage, S.D.D.; Dawe, A.L.; Wang, L.; Wang, Y. Potato spindle tuber viroid modulates its replication through a direct interaction with a splicing regulator. J. Virol. 2018, 92, e01004-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudiyanselage, S.D.D.; Qu, J.; Tian, N.; Jiang, J.; Wang, Y. Potato spindle tuber viroid RNA-templated transcription: Factors and regulation. Viruses 2018, 10, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matoušek, J.; Steinbachová, L.; Drábková, L.Z.; Kocábek, T.; Potěšil, D.; Mishra, A.K.; Honys, D. Elimination of viroids from tobacco pollen involves a decrease in propagation rate and an increase of the degradation processes. Int. J. Mol. Sci. 2020, 21, 3029. [Google Scholar] [CrossRef]
- Matoušek, J.; Patzak, J. A low transmissibility of hop latent viroid through a generative phase of Humulus lupulus L. Biol. Plant. 2000, 43, 145–148. [Google Scholar] [CrossRef]
- Matoušek, J.; Kocábek, T.; Mishra, A.K.; Radišek, S.; Steger, G. The decrease of the propagation rate of hop viroids in pollen and a potential multifunctional role of transcription factor TFIIIA. In Book of Abstracts, V. International Humulus Symposium; University of Hohenheim: Stuttgart, Germany, 8−12 March 2021; p. 36. [Google Scholar]
- Patzak, J.; Krofta, K.; Henychová, A.; Nesvadba, V. Number and size of lupulin glands, glandular trichomes of hop (Humulus lupulus L.), play a key role in contents of bitter acids and polyphenols in hop cone. Int. J. Food Sci. Technol. 2015, 50, 1864–1872. [Google Scholar] [CrossRef]
- Krofta, K. Contents of xanthohumol in Czech hops. Kvasný Prům. 2003, 49, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Krofta, K.; Poustka, J.; Nováková, K.; Hajšlová, J. Contents of prenylflavonoids in Czech hops and beers. Acta Hortic. 2005, 668, 201–206. [Google Scholar] [CrossRef]
- Nesvadba, V.; Krofta, K. Variability in the contents of important compounds for pharmaceutical and brewing industries within hop gene pool. Agriculture 2009, 55, 10–16. [Google Scholar]
- Patzak, J.; Henychová, A.; Matoušek, J. Developmental regulation of lupulin gland-associated genes in aroma and bitter hops (Humulus lupulus L.). BMC Plant Biol. 2021. [Google Scholar]
- Samanta, A.; Das, G.; Das, S.K. Roles of flavonoids in plants. Int. J. Pharm. Sci. Technol. 2011, 100, 12–35. [Google Scholar]
- Feiner, A.; Pitra, N.; Matthews, P.; Pillen, K.; Wessjohann, L.A.; Riewe, D. Downy mildew resistance is genetically mediated by prophylactic production of phenylpropanoids in hop. Plant Cell Environ. 2021, 44, 323–338. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, R.A. Plant terpenes: Defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Patzak, J.; Henychová, A.; Svoboda, P.; Malířová, I. Assessment of epigenetic methylation changes in hop (Humulus lupulus) plants obtained by meristem culture. Czech J. Genet. Plant Breed. 2020, 56, 159–164. [Google Scholar] [CrossRef]
- Patzak, J. Assessment of somaclonal variability in hop (Humulus lupulus L.) in vitro meristem cultures and clones by molecular methods. Euphytica 2003, 131, 343–350. [Google Scholar] [CrossRef]
- Patzak, J.; Svoboda, P.; Henychová, A.; Malířová, I. Detection of hop viruses and viroids by qRT-PCR in the Czech Republic. In Proceedings of the Scientific Commission of IHGC, Stefan am Walde, Austria, 25–29 June 2017; pp. 101–3785. [Google Scholar]
- Patzak, J.; Nesvadba, V.; Krofta, K.; Henychová, A.; Marzoev, A.I.; Richards, K. Evaluation of genetic variability of wild hops (Humulus lupulus L.) in Canada and the Caucasus region by chemical and molecular methods. Genome 2010, 53, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Natsume, S.; Takagi, H.; Shiraishi, A.; Murata, J.; Toyonaga, H.; Patzak, J.; Takagi, M.; Yaegashi, H.; Uemura, A.; Mitsuoka, C.; et al. The draft genome of hop (Humulus lupulus), an essence for brewing. Plant Cell Physiol. 2015, 56, 428–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, S.T.; Sudarsanam, R.; Henning, J.; Hendrix, D. HopBase: A unified resource for Humulus genomics. Database 2017, 2017, bax009. [Google Scholar] [CrossRef] [Green Version]
- Padgitt-Cobb, L.K.; Kingan, S.B.; Wells, J.; Elser, J.; Kronmiller, B.; Moore, D.; Concepcion, G.; Peluso, P.; Rank, D.; Jaiswal, P.; et al. A draft phased assembly of the diploid Cascade hop (Humulus lupulus) genome. Plant Genome 2021, 14, e20072. [Google Scholar] [CrossRef] [PubMed]






| HLVd Infection | Negative | Positive | Negative | Positive |
|---|---|---|---|---|
| Harvest year | 2019 | 2019 | 2020 | 2020 |
| Number of samples | 28 | 41 | 14 | 56 |
| Alpha acids (% of DW) | 5.54 ± 0.63 | 3.84 ± 0.72 *** | 5.46 ± 0.99 | 3.59 ± 0.60 *** |
| Beta acids (% of DW) | 3.78 ± 0.32 | 3.96 ± 0.47 * | 3.57 ± 0.34 | 3.63 ± 0.31 |
| Cohumulone (% of AA) | 21.34 ± 2.11 | 21.86 ± 2.50 | 20.07 ± 1.13 | 21.47 ± 2.13 ** |
| Colupulone (% of BA) | 43.07 ± 1.92 | 41.45 ± 3.07 ** | 38.79 ± 0.90 | 39.22 ± 2.28 |
| Xanthohumol (% of DW) | 0.309 ± 0.035 | 0.242 ± 0.040 *** | 0.289 ± 0.032 | 0.221 ± 0.034 *** |
| Cultivar | Sládek | Premiant | Agnus | |||
|---|---|---|---|---|---|---|
| HLVd infection | Negative | Positive | Negative | Positive | Negative | Positive |
| Number of samples | 9 | 7 | 3 | 3 | 3 | 4 |
| Alpha acids (% of DW) | 9.31 ± 0.98 | 6.68 ± 0.17 *** | 8.86 ± 0.30 | 7.55 ± 0.34 *** | 11.53 ± 0.45 | 10.52 ± 0.27 ** |
| Beta acids (% of DW) | 3.83 ± 0.46 | 4.44 ± 0.24 *** | 3.32 ± 0.41 | 3.23 ± 0.33 | 3.90 ± 0.28 | 3.52 ± 0.20 * |
| Cohumulone (% of AA) | 25.31 ± 0.55 | 23.63 ± 0.39 *** | 17.13 ± 1.62 | 16.33 ± 0.51 | 29.97 ± 1.08 | 30.55 ± 0.61 |
| Colupulone (% of BA) | 49.21 ± 0.82 | 49.29 ± 0.47 | 37.30 ± 3.32 | 38.23 ± 2.12 | 52.80 ± 1.31 | 53.50 ± 0.57 |
| Xanthohumol (% of DW) | 0.606 ± 0.045 | 0.467 ± 0.027 *** | 0.313 ± 0.021 | 0.247 ± 0.006 *** | 0.747 ± 0.067 | 0.718 ± 0.058 |
| HLVd Infection | Negative | Positive | Negative | Positive |
|---|---|---|---|---|
| Harvest year | 2019 | 2019 | 2020 | 2020 |
| Number of samples | 21 | 6 | 14 | 56 |
| Total oils (% of DW) | 0.527 ± 0.146 | 0.438 ± 0.108 | 0.443 ± 0.094 | 0.481 ± 0.112 |
| Monoterpenes (% of EO) | ||||
| Myrcene | 19.75 ± 3.12 | 24.60 ± 5.82 ** | 22.79 ± 3.71 | 25.26 ± 3.81 ** |
| α-pinene | 0.053 ± 0.013 | 0.083 ± 0.027 *** | 0.082 ± 0.025 | 0.116 ± 0.026 *** |
| β-pinene | 0.414 ± 0.070 | 0.603 ± 0.157 *** | 0.615 ± 0.161 | 0.871 ± 0.186 *** |
| Sesquiterpenes (% of EO) | ||||
| α-humulene | 20.72 ± 2.52 | 19.08 ± 2.27 | 20.38 ± 2.59 | 20.17 ± 2.67 |
| β-caryophyllene | 8.74 ± 1.00 | 7.02 ± 1.58 *** | 7.09 ± 1.11 | 6.68 ± 1.03 |
| β-farnesene | 23.50 ± 3.00 | 20.79 ± 3.38 * | 22.13 ± 3.00 | 17.96 ± 2.82 *** |
| γ-muurolene | 1.098 ± 0.165 | 0.892 ± 0.263 ** | 0.796 ± 0.106 | 0.765 ± 0.120 |
| β-bisabolene | 0.409 ± 0.097 | 0.420 ± 0.040 | 0.474 ± 0.061 | 0.405 ± 0.093 *** |
| γ-cadinene | 1.136 ± 0.172 | 0.912 ± 0.267 ** | 0.806 ± 0.107 | 0.779 ± 0.130 |
| δ-cadinene | 1.892 ± 0.257 | 1.512 ± 0.404 *** | 1.295 ± 0.204 | 1.214 ± 0.201 |
| Selinenes | 1.626 ± 0.462 | 1.588 ± 1.086 | 1.108 ± 0.168 | 1.064 ± 0.244 |
| Alcohols (% of EO) | ||||
| Linalool | 0.201 ± 0.064 | 0.303 ± 0.084 *** | 0.337 ± 0.124 | 0.554 ± 0.175 *** |
| Geraniol | 0.035 ± 0.014 | 0.218 ± 0.127 *** | 0.204 ± 0.278 | 0.514 ± 0.272 *** |
| Epoxides A (% of EO) | 0.859 ± 0.483 | 0.995 ± 0.397 | 1.558 ± 0.522 | 3.070 ± 1.173 *** |
| Ketones B (% of EO) | 2.637 ± 0.461 | 2.302 ± 0.289 | 2.838 ± 0.495 | 1.970 ± 0.483 *** |
| Esters (% of EO) | ||||
| Methylgeranate | 0.082 ± 0.035 | 0.348 ± 0.150 *** | 0.198 ± 0.139 | 0.672 ± 0.381 *** |
| Methylheptanoate | 0.261 ± 0.087 | 0.259 ± 0.078 | 0.329 ± 0.090 | 0.434 ± 0.109 *** |
| Methyloctanoate | 0.547 ± 0.204 | 0.432 ± 0.063 | 0.420 ± 0.163 | 0.314 ± 0.132 ** |
| Methylnon-6-enoate | 0.069 ± 0.030 | 0.093 ± 0.056 | 0.084 ± 0.035 | 0.157 ± 0.029 *** |
| Methyl-8-methylnonanoate | 0.158 ± 0.053 | 0.121 ± 0.043 | 0.163 ± 0.046 | 0.114 ± 0.043 *** |
| Methyldeca-4,8-dienoate | 0.433 ± 0.118 | 0.485 ± 0.113 | 0.409 ± 0.121 | 0.702 ± 0.126 *** |
| Methyldecanoate | 0.383 ± 0.137 | 0.288 ± 0.045 | 0.259 ± 0.096 | 0.140 ± 0.081 *** |
| Methyldodeca-3,6-dienoate | 0.924 ± 0.180 | 0.827 ± 0.070 | 0.813 ± 0.146 | 0.550 ± 0.173 *** |
| Cultivar | Sládek | Premiant | Agnus | |||
|---|---|---|---|---|---|---|
| HLVd infection | Negative | Positive | Negative | Positive | Negative | Positive |
| Number of samples | 7 | 4 | 2 | 2 | 2 | 2 |
| Total oils (% of DW) | 1.836 ± 0.403 | 1.385 ± 0.130 * | 1.270 ± 0481 | 1.085 ± 0.417 | 2.005 ± 0.813 | 1.775 ± 0.488 |
| Monoterpenes (% of EO) | ||||||
| Myrcene | 34.06 ± 1.68 | 30.05 ± 5.60 | 29.65 ± 9.69 | 28.95 ± 4.03 | 32.35 ± 5.59 | 32.20 ± 5.37 |
| α-pinene | 0.087 ± 0.013 | 0.098 ± 0.025 | 0.070 ± 0.014 | 0.090 ± 0 | 0.165 ± 0.050 | 0.195 ± 0.007 |
| β-pinene | 0.677 ± 0.024 | 0.798 ± 0.139 ** | 0.595 ± 0.233 | 0.720 ± 0.113 | 1.055 ± 0.007 | 1.275 ± 0.304 |
| Sesquiterpenes (% of EO) | ||||||
| α-humulene | 29.73 ± 1.27 | 31.50 ± 4.37 | 33.10 ± 7.21 | 33.65 ± 7.57 | 22.50 ± 0.57 | 20.55 ± 2.19 |
| β-caryophyllene | 12.20 ± 0.64 | 13.18 ± 2.15 | 10.90 ± 1.70 | 10.76 ± 1.48 | 13.80 ± 0.42 | 13.10 ± 1.41 |
| β-farnesene | 0.103 ± 0.119 | 0.093 ± 0.085 | 2.540 ± 0.255 | 1.885 ± 0.714 | 0.280 ± 0.071 | 0.185 ± 0.064 |
| γ-muurolene | 1.120 ± 0.180 | 1.113 ± 0.147 | 0.925 ± 0.085 | 0.800 ± 0 | 1.180 ± 0.170 | 1.220 ± 0.130 |
| β-bisabolene | 0.201 ± 0.172 | 0.048 ± 0.082 | 0.060 ± 0.060 | 0.165 ± 0.165 | 0 | 0 |
| γ-cadinene | 1.077 ± 0.039 | 1.153 ± 0.172 | 0.975 ± 0.105 | 1.153 ± 0.172 | 1.235 ± 0.145 | 1.250 ± 0.130 |
| δ-cadinene | 1.864 ± 0.109 | 2.018 ± 0.311 | 1.705 ± 0.205 | 1.570 ± 0.080 | 1.910 ± 0.170 | 1.830 ± 0.250 |
| Selinenes | 1.240 ± 0.247 | 1.158 ± 0.400 | 1.440 ± 0.240 | 1.100 ± 0.198 | 2.910 ± 0.655 | 2.940 ± 0.622 |
| Alcohols (% of EO) | ||||||
| Linalool | 0.240 ± 0.028 | 0.310 ± 0.075 ** | 0.605 ± 0.262 | 0.600 ± 0.035 | 0.570 ± 0.113 | 0.625 ± 0.035 |
| Geraniol | 0.200 ± 0.064 | 0.343 ± 0.056 *** | 0.050 ± 0.028 | 0.175 ± 0.021 ** | 0.740 ± 0.311 | 0.730 ± 0.325 |
| EpoxidesA (% of EO) | 0.547 ± 0.396 | 1.178 ± 0.553 * | 0.280 ± 0.028 | 0.235 ± 0.035 | 2.315 ± 0.813 | 3.140 ± 0.354 |
| KetonesB (% of EO) | 2.934 ± 0.632 | 2.330 ± 0.756 | 3.325 ± 0.813 | 1.990 ± 0.057 | 1.120 ± 0.057 | 1.180 ± 0.141 |
| Esters (% of EO) | ||||||
| Methylgeranate | 0.330 ± 0.147 | 1.030 ± 0.301 *** | 0.198 ± 0.139 | 0.405 ± 0.163 | 2.480 ± 0.368 | 2.950 ± 0.297 |
| Methylheptanoate | 0.407 ± 0.067 | 0.330 ± 0.149 | 0.395 ± 0.064 | 0.355 ± 0.064 | 0.190 ± 0.028 | 0.225 ± 0.212 |
| Methyloctanoate | 0.837 ± 0.238 | 0.618 ± 0.303 | 0.315 ± 0.021 | 0.295 ± 0.177 | 0.225 ± 0.021 | 0.260 ± 0.071 |
| Methylnon-6-enoate | 0.044 ± 0.013 | 0.058 ± 0.015 | 0.060 ± 0.028 | 0.080 ± 0.028 | 0.040 ± 0.014 | 0.040 ± 0.014 |
| Methyl-8-methylnonanoate | 0.084 ± 0.026 | 0.065 ± 0.033 | 0.110 ± 0.014 | 0.060 ± 0 ** | 0.195 ± 0.007 | 0.200 ± 0.014 |
| Methyldeca-4,8-dienoate | 0.513 ± 0.089 | 0.628 ± 0.213 | 0.570 ± 0.255 | 0.865 ± 0.431 | 0.340 ± 0.057 | 0.390 ± 0.127 |
| Methyldecanoate | 0.410 ± 0.127 | 0.270 ± 0.123 | 0.245 ± 0.021 | 0.160 ± 0.071 | 0.125 ± 0.035 | 0.150 ± 0.071 |
| Methyldodeca-3,6-dienoate | 0.390 ± 0.288 | 0.145 ± 0.290 | 0.285 ± 0.403 | 0.265 ± 0.375 | 0 | 0 |
| Abbrev. | Gene | Number * |
|---|---|---|
| Bitter acids biosynthesis | ||
| BCAT1 | Branched-chain amino acid aminotransferase 1 | 002627F.g2, JQ063073 |
| VPS | Phloroisovalerophenone synthase | 001329F.g74, FJ554588 |
| PT1L | 2-acylphloroglucinol 4-prenyltransferase | KM222441 |
| PT2 | 2-acyl-4-prenylphloroglucinol 6-prenyltransferase | KM222442 |
| HS1 | Monooxygenase 2 (Humulone synthase 1) | 010625F.g1, 008956F.g7, KJ398144 |
| HS2 | Monooxygenase 2 (Humulone synthase 2) | 008118F.g14, KJ398145 |
| Polyphenols and flavonoids biosynthesis | ||
| Flavonoids biosynthesis | ||
| PAL | Phenylalanine ammonia-lyase | 000198F.g108, g113, g114, g115, g116, g117 |
| 4CL2 | 4-coumarate-CoA ligase 2 | 001395F.g25 |
| CHSH1 | Naringenin-chalcone synthase (CHS_H1) | 000203F.g58, AJ304877 |
| OMT1 | O-methyltransferase 1 (Desmethylxanthohumol 6’-O-methyltransferase) | 000009F.g116, EU309725 |
| F3H | Flavanone 3-hydroxylase | 001909F.g33 |
| LAR | Leucoanthocyanidin reductase | 000109F.g58, HQ734722 |
| Terpenes biosynthesis | ||
| GPPS-SSU | Geranyl diphosphate synthase small subunit | 001483F.g7, FJ455406 |
| FPPS | Farnesyl pyrophosphate synthase | 000817F.g23, AB053487, AF268889 |
| MTS1 | Monoterpene synthase 1 | 000149F.g29, EU760348 |
| MTS2 | Monoterpene synthase 2 (myrcene synthase) | 003722F.g32, EU760349 |
| STS1 | Sesquiterpene synthase 1 (α-humulene synthase) | 001011F.g31, EU760350 |
| STS2 | Sesquiterpene synthase 2 (germacrene-A synthase) | 001011F.g31, EU760351 |
| TPS9 | Terpene synthase 9 | 001370F.g12 |
| NES | (E)-nerolidol/linalool synthase | 004063F.g13 |
| Transcription regulation of biosynthesis genes | ||
| MYB3 | Transcription factor HlMYB3 | AM501509 |
| MYB8 | Transcription factor HlMYB8 | 002031F.g25, HG983335 |
| MYB78 | Transcription factor MYB78 | 000063F.g63 |
| bHLH2 | Transcription factor HlbHLH2 (TT8) | 000662F.g4, FR751553 |
| bHLH4 | Transcription factor GLABRA 3 (HlbHLH4) | 001145F.g21, HG983336 |
| WRKY1 | WRKY transcription factor 1 | 000029F.g2, CBY88801 |
| TFIIIA | Transcription factor IIIA | 002165F.g6 |
| RPL5 | Ribosomal protein L5 | GAAW01025872 |
| Reference genes | ||
| TTG1 | Protein TRANSPARENT TESTA GLABRA 1 (HlWD40) | 002162F.g15, FN689721 |
| MYC2 | Transcription factor MYC2 | 001862F.g5 |
| PIF4 | Transcription factor PIF4 | 000802F.g1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 004935F.g2, 004935F.g5 |
| RH46 | DEAD-box ATP-dependent RNA helicase 46 | 000004F.g75 |
| cis elements | MYB1AT | MYB2CONSENSUSAT | MYBCORE | MYB1LEPR | MYBCOREATCYCB1 | MYBPZM | BOXLCOREDCPAL | MYBPLANT | MYBST1 | MYBGAHV | MYCCONSENSUSAT | WRKY71OS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence/gene | WAACCA | YAACKG | CNGTTR | GTTAGTT | AACGG | CCWACC | ACCWWCC | MACCWAMC | GGATA | TAACAAA | CANNTG | TGAC |
| BCAT1 | −439 | −1688 | −1745, −1089, −925, −792, −695 | −1643, −1093, −890, −584 | ||||||||
| VPS | −1559 | −1924, −1914, −1826, −1733, −1715, −1702, −474 | −1873, −1236, −874 | −1865 | −1845, −1074 | −1554 | −1555 | −1556, −1473, −1431 | −1020 | −996 | −1924, −1826, −1770, −1733, −1702, −1488, −1448, −1316, −811, −474, −77 | −1920, −1886, −1852, −1815, −1689, −1679, −1676, −961, −470 |
| PT1L | −512 | −1536, −1313 | −1334, −1153 | −1801, −401, −397, −205 | −398 | −399 | −1636, −1488, −769 | −1106 | −1611, −1546, −1408, −1325, −333 | −1987, −1698, −1507, −1420, −1057, −980, −928, −638, −597, −558 | ||
| PT2 | −466 | −1810, −1331, −271 | −27 | −902, −350, −271, −8 | −1891, −1634, −1001, −777, −510, −345, −340 | |||||||
| HS1 | −126 | −1907 | −1905 | −953 | −431 | −432 | −433 | −1609, −1526, −1509, −1433, 537, −490, −425, −224, −187 | −1959, −1917, −1860, −647, −594, −495, −280 | |||
| HS2 | −120 | −349 | −1612 | −137 | −1809, −1730, −1714, 1624, −1423, −600, −538 | −921, −814, −734, −605, −427, −398, −345, −243 | ||||||
| PAL | −830, −539, −391, −321, −143, −116 | −1242, −665, −308 | −919, −492 | −493 | −494 | −1450 | −1998, −789 | −1979, −1727, −1259, −1211, −894 | −1648, −1265, −1024, −835, −577, −414, −409, −333, −237, −233, −176 | |||
| 4CL2 | −937, −726, −361 | −1896, −1790 | −1877, −1694, −183 | −169, −118 | −170, −119 | −1238, −1112 | −1146 | −694, −547, −497, −300, −144 | −1438, −1011, −800 | |||
| CHSH1 | −1800 | −837, −476 | −1526, −385 | −622 | −836, −475 | −1427, −517 | −1861 | −245 | −1303, −1042, −224 | −1665, −1640, −1283, 1179, 1148, −725, −284, −220 | ||
| OMT1 | −1769, −556 | −1496, −1306 | −1034, −393 | |||||||||
| F3H | −1499, −141, −134, −110 | −1441 | −1544, −210 | −1934, −1538, −1468, −881, −734, −632, −397 | −1937, −1634, −1534, −1277, −330, −256 | |||||||
| LAR | −978, −390 | −1948, −1711, −1580 | −1578 | −1723, −297 | −1967, −1877, −1267, −974, −830, −82 | −650, −609, −328 | ||||||
| GPPS-SSU | −1905, −1870, −1788, −1515, −1222, −270, −8 | −1984, −30 | −1750 | −635 | −1736, −1296 | −1297 | −1787, −1298 | −748 | −625 | −1987, −1766, −1495, −1344, −727, −571, −366, −305, −189, −49 | −1947, −1693, −1491, −805, −723 | |
| FPPS | −26 | −1842, −1092, −772, −404, −174 | −1779, −1698, −1661, −1337, −595, −119 | |||||||||
| MTS1 | −1305 | −193, −173 | −1305, −1098 | −1253, −845, −386, −117 | −1930 | |||||||
| MTS2 | −905, −566 | −1796 | −1193 | −1993 | −1194 | −1933, −399 | −1407, −1345, −978, −836, −708, −657, −536, −312 | |||||
| STS1 | −1196, −455 | −1875 | −983 | −810, −733, −531, −471, −65 | −1407, −1090, −640, −527, −498, −297 | |||||||
| STS2 | −1212, −452 | −1892 | −995 | −807, −730, −528, −468, −65 | −1423, −1105, −637, −524, −495, −294 | |||||||
| TPS9 | −1909, −283 | −1160, −12 | −18 | −1273, −585, −12 | −1814, −1555, −1305, −1257 | |||||||
| NES | −618 | −929, −692 | −545, −500, −327 | −928 | −1411 | −834 | −1634 | −1865, −1796, −1539, −1476, −1101, −692, −500, −462, −310 | −1286, −1210, −788, −688, −313, −100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patzak, J.; Henychová, A.; Krofta, K.; Svoboda, P.; Malířová, I. The Influence of Hop Latent Viroid (HLVd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus lupulus L.) Glandular Trichomes. Plants 2021, 10, 2297. https://doi.org/10.3390/plants10112297
Patzak J, Henychová A, Krofta K, Svoboda P, Malířová I. The Influence of Hop Latent Viroid (HLVd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus lupulus L.) Glandular Trichomes. Plants. 2021; 10(11):2297. https://doi.org/10.3390/plants10112297
Chicago/Turabian StylePatzak, Josef, Alena Henychová, Karel Krofta, Petr Svoboda, and Ivana Malířová. 2021. "The Influence of Hop Latent Viroid (HLVd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus lupulus L.) Glandular Trichomes" Plants 10, no. 11: 2297. https://doi.org/10.3390/plants10112297
APA StylePatzak, J., Henychová, A., Krofta, K., Svoboda, P., & Malířová, I. (2021). The Influence of Hop Latent Viroid (HLVd) Infection on Gene Expression and Secondary Metabolite Contents in Hop (Humulus lupulus L.) Glandular Trichomes. Plants, 10(11), 2297. https://doi.org/10.3390/plants10112297





