New Insights into Plant TPK Ion Channel Evolution
Abstract
:1. Introduction
2. Results
2.1. Analysis of the Arabidopsis C- and N-Terminal Pore Regions
2.2. Pore Alignment and Analysis of the Voltage-Dependent Potassium Channel Signature
2.3. Variation in the Selectivity Filter Motif in the First and Second Pore
2.4. TPK/KCO Phylogeny
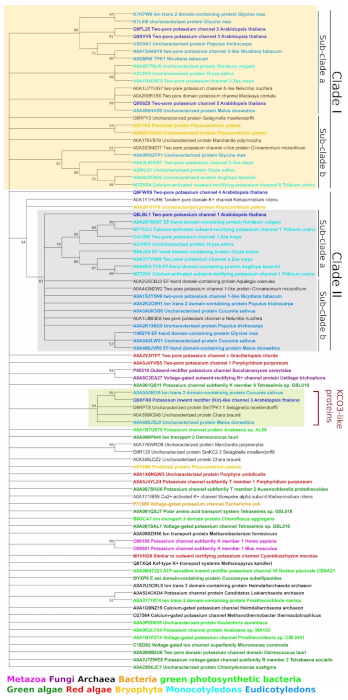
2.4.1. General TPK Phylogeny (Based on the Alignment of Full-Length Proteins)
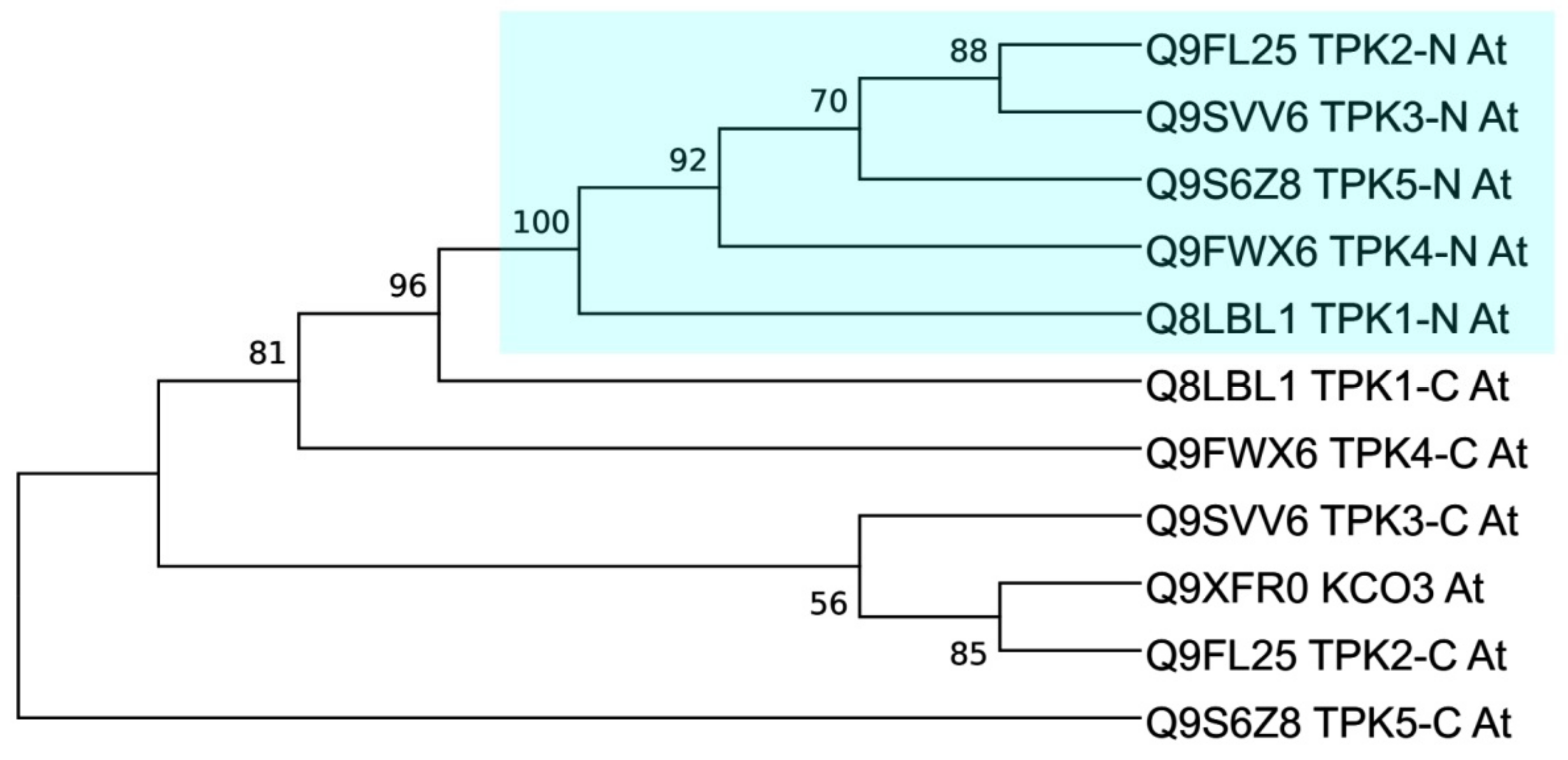
2.4.2. Domain-Based TPK Phylogeny (First and Second Pores Taken Separately)
3. Discussion
4. Materials and Methods
4.1. Identification of the TPK Family Members
4.2. Multiple Sequence Alignments and Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maathuis, F.J. Physiological Functions of Mineral Macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Marschner, P. (Ed.) Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Elsevier: London, UK, 2012; ISBN 978-0-12-384905-2. [Google Scholar]
- Britto, D.T.; Coskun, D.; Kronzucker, H.J. Potassium Physiology from Archean to Holocene: A Higher-Plant Perspective. J. Plant Physiol. 2021, 262, 153432. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of Potassium Transport in Plants under Hostile Conditions: Implications for Abiotic and Biotic Stress Tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, X.; Giraldo, J.P.; Shabala, S. It Is Not All about Sodium: Revealing Tissue Specificity and Signalling Roles of Potassium in Plant Responses to Salt Stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The Effect of Potassium Nutrition on Pest and Disease Resistance in Plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Nanatani, K.; Hamamoto, S.; Shimizu, M.; Takahashi, M.; Tabuchi-Kobayashi, M.; Mizutani, A.; Schroeder, J.I.; Souma, S.; Uozumi, N. Defining Membrane Spanning Domains and Crucial Membrane-Localized Acidic Amino Acid Residues for K+ Transport of a Kup/HAK/KT-Type Escherichia coli Potassium Transporter. J. Biochem. 2014, 155, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Isayenkov, S.V.; Dabravolski, S.A.; Pan, T.; Shabala, S. Phylogenetic Diversity and Physiological Roles of Plant Monovalent Cation/H+ Antiporters. Front. Plant Sci. 2020, 11, 573564. [Google Scholar] [CrossRef] [PubMed]
- Lebaudy, A.; Véry, A.-A.; Sentenac, H. K+ Channel Activity in Plants: Genes, Regulations and Functions. FEBS Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Maathuis, F.J.M. Physiological Roles of Nonselective Cation Channels in Plants: From Salt Stress to Signalling and Development. New Phytol. 2007, 175, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Dreyer, I.; Riedelsberger, J. The Role of K+ Channels in Uptake and Redistribution of Potassium in the Model Plant Arabidopsis Thaliana. Front. Plant Sci. 2013, 4, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bihler, H.; Eing, C.; Hebeisen, S.; Roller, A.; Czempinski, K.; Bertl, A. TPK1 Is a Vacuolar Ion Channel Different from the Slow-Vacuolar Cation Channel. Plant Physiol. 2005, 139, 417–424. [Google Scholar] [CrossRef] [Green Version]
- Gobert, A.; Isayenkov, S.; Voelker, C.; Czempinski, K.; Maathuis, F.J.M. The Two-Pore Channel TPK1 Gene Encodes the Vacuolar K+ Conductance and Plays a Role in K+ Homeostasis. Proc. Natl. Acad. Sci. USA 2007, 104, 10726–10731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latz, A.; Ivashikina, N.; Fischer, S.; Ache, P.; Sano, T.; Becker, D.; Deeken, R.; Hedrich, R. In Planta AKT2 Subunits Constitute a PH- and Ca2+-Sensitive Inward Rectifying K+ Channel. Planta 2007, 225, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Hartley, T.N.; Maathuis, F.J.M. Allelic Variation in the Vacuolar TPK1 Channel Affects Its Calcium Dependence and May Impact on Stomatal Conductance. FEBS Lett. 2016, 590, 110–117. [Google Scholar] [CrossRef]
- Isner, J.C.; Begum, A.; Nuehse, T.; Hetherington, A.M.; Maathuis, F.J.M. KIN7 Kinase Regulates the Vacuolar TPK1 K+ Channel during Stomatal Closure. Curr. Biol. 2018, 28, 466–472.e4. [Google Scholar] [CrossRef] [Green Version]
- Latz, A.; Becker, D.; Hekman, M.; Müller, T.; Beyhl, D.; Marten, I.; Eing, C.; Fischer, A.; Dunkel, M.; Bertl, A.; et al. TPK1, a Ca2+-Regulated Arabidopsis Vacuole Two-Pore K+ Channel Is Activated by 14-3-3 Proteins. Plant J. 2007, 52, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Voelker, C.; Gomez-Porras, J.L.; Becker, D.; Hamamoto, S.; Uozumi, N.; Gambale, F.; Mueller-Roeber, B.; Czempinski, K.; Dreyer, I. Roles of Tandem-Pore K+ Channels in Plants—A Puzzle Still to Be Solved. Plant Biol. 2010, 12, 56–63. [Google Scholar] [CrossRef]
- Tang, R.-J.; Wang, C.; Li, K.; Luan, S. The CBL–CIPK Calcium Signaling Network: Unified Paradigm from 20 Years of Discoveries. Trends Plant Sci. 2020, 25, 604–617. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Bai, B.; Almutairi, B.O.; Kudla, J. Emerging Roles of the CBL-CIPK Calcium Signaling Network as Key Regulatory Hub in Plant Nutrition. J. Plant Physiol. 2021, 257, 153335. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M. Vacuolar Two-Pore K+ Channels Act as Vacuolar Osmosensors. New Phytol. 2011, 191, 84–91. [Google Scholar] [CrossRef]
- Voelker, C.; Schmidt, D.; Mueller-Roeber, B.; Czempinski, K. Members of the Arabidopsis AtTPK/KCO Family Form Homomeric Vacuolar Channels in Planta. Plant J. 2006, 48, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Dunkel, M.; Latz, A.; Schumacher, K.; Müller, T.; Becker, D.; Hedrich, R. Targeting of Vacuolar Membrane Localized Members of the TPK Channel Family. Mol. Plant 2008, 1, 938–949. [Google Scholar] [CrossRef]
- Marcel, D.; Müller, T.; Hedrich, R.; Geiger, D. K+ Transport Characteristics of the Plasma Membrane Tandem-Pore Channel TPK4 and Pore Chimeras with Its Vacuolar Homologs. FEBS Lett. 2010, 584, 2433–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isayenkov, S.; Isner, J.-C.; Maathuis, F.J.M. Rice Two-Pore K+ Channels Are Expressed in Different Types of Vacuoles. Plant Cell 2011, 23, 756–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isayenkov, S.; Isner, J.-C.; Maathuis, F.J.M. Membrane Localisation Diversity of TPK Channels and Their Physiological Role. Plant Signal. Behav. 2011, 6, 1201–1204. [Google Scholar] [CrossRef] [Green Version]
- Becker, D.; Geiger, D.; Dunkel, M.; Roller, A.; Bertl, A.; Latz, A.; Carpaneto, A.; Dietrich, P.; Roelfsema, M.R.G.; Voelker, C.; et al. AtTPK4, an Arabidopsis Tandem-Pore K+ Channel, Poised to Control the Pollen Membrane Voltage in a PH- and Ca2+-Dependent Manner. Proc. Natl. Acad. Sci. USA 2004, 101, 15621–15626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höhner, R.; Galvis, V.C.; Strand, D.D.; Völkner, C.; Krämer, M.; Messer, M.; Dinc, F.; Sjuts, I.; Bölter, B.; Kramer, D.M.; et al. Photosynthesis in Arabidopsis Is Unaffected by the Function of the Vacuolar K+ Channel TPK3. Plant Physiol. 2019, 180, 1322–1335. [Google Scholar] [CrossRef] [Green Version]
- Maîtrejean, M.; Wudick, M.M.; Voelker, C.; Prinsi, B.; Mueller-Roeber, B.; Czempinski, K.; Pedrazzini, E.; Vitale, A. Assembly and Sorting of the Tonoplast Potassium Channel AtTPK1 and Its Turnover by Internalization into the Vacuole. Plant Physiol. 2011, 156, 1783–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaślan, D.; Dreyer, I.; Lu, J.; O’Malley, R.; Dindas, J.; Marten, I.; Hedrich, R. Voltage-Dependent Gating of SV Channel TPC1 Confers Vacuole Excitability. Nat. Commun. 2019, 10, 2659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocchetti, A.; Sharma, T.; Wulfetange, C.; Scholz-Starke, J.; Grippa, A.; Carpaneto, A.; Dreyer, I.; Vitale, A.; Czempinski, K.; Pedrazzini, E. The Putative K+ Channel Subunit AtKCO3 Forms Stable Dimers in Arabidopsis. Front. Plant Sci. 2012, 3, 251. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.M.; Schroeder, J.I. Calcium-Activated K+ Channels and Calcium-Induced Calcium Release by Slow Vacuolar Ion Channels in Guard Cell Vacuoles Implicated in the Control of Stomatal Closure. Plant Cell 1994, 6, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Porras, J.L.; Riaño-Pachón, D.M.; Benito, B.; Haro, R.; Sklodowski, K.; Rodríguez-Navarro, A.; Dreyer, I. Phylogenetic Analysis of K+ Transporters in Bryophytes, Lycophytes, and Flowering Plants Indicates a Specialization of Vascular Plants. Front. Plant Sci. 2012, 3, 167. [Google Scholar] [CrossRef] [Green Version]
- Uehara, C.; Takeda, K.; Ibuki, T.; Furuta, T.; Hoshi, N.; Tanudjaja, E.; Uozumi, N. Analysis of Arabidopsis TPK2 and KCO3 Reveals Structural Properties Required for K+ Channel Function. Channels 2020, 14, 336–346. [Google Scholar] [CrossRef]
- Gutman, G.A.; Chandy, K.G.; Grissmer, S.; Lazdunski, M.; Mckinnon, D.; Pardo, L.A.; Robertson, G.A.; Rudy, B.; Sanguinetti, M.C.; Stühmer, W.; et al. International Union of Pharmacology. LIII. Nomenclature and Molecular Relationships of Voltage-Gated Potassium Channels. Pharmacol. Rev. 2005, 57, 473–508. [Google Scholar] [CrossRef]
- Jegla, T.; Busey, G.; Assmann, S.M. Evolution and Structural Characteristics of Plant Voltage-Gated K+ Channels. Plant Cell 2018, 30, 2898–2909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, M.L.; Krovetz, H.S.; VanDongen, A.M.J. GYGD Pore Motifs in Neighbouring Potassium Channel Subunits Interact to Determine Ion Selectivity. J. Physiol. 2001, 530, 21–33. [Google Scholar] [CrossRef]
- Hamamoto, S.; Marui, J.; Matsuoka, K.; Higashi, K.; Igarashi, K.; Nakagawa, T.; Kuroda, T.; Mori, Y.; Murata, Y.; Nakanishi, Y.; et al. Characterization of a Tobacco TPK-Type K+ Channel as a Novel Tonoplast K+ Channel Using Yeast Tonoplasts. J. Biol. Chem. 2008, 283, 1911–1920. [Google Scholar] [CrossRef] [Green Version]
- Ivashikina, N.; Hedrich, R. K+ Currents through SV-Type Vacuolar Channels Are Sensitive to Elevated Luminal Sodium Levels: Sodium-Sensitive SV Channels. Plant J. 2005, 41, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Moniruzzaman, M.; Weinheimer, A.R.; Martinez-Gutierrez, C.A.; Aylward, F.O. Widespread Endogenization of Giant Viruses Shapes Genomes of Green Algae. Nature 2020, 588, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Finke, J.; Winget, D.; Chan, A.; Suttle, C. Variation in the Genetic Repertoire of Viruses Infecting Micromonas Pusilla Reflects Horizontal Gene Transfer and Links to Their Environmental Distribution. Viruses 2017, 9, 116. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Husnik, F.; McCutcheon, J.P. Functional Horizontal Gene Transfer from Bacteria to Eukaryotes. Nat. Rev. Microbiol. 2018, 16, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fu, Y.; Jiang, D.; Li, G.; Xie, J.; Cheng, J.; Peng, Y.; Ghabrial, S.A.; Yi, X. Widespread Horizontal Gene Transfer from Double-Stranded RNA Viruses to Eukaryotic Nuclear Genomes. J. Virol. 2010, 84, 11876–11887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam Protein Families Database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE Team. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabravolski, S.A.; Isayenkov, S.V. New Insights into Plant TPK Ion Channel Evolution. Plants 2021, 10, 2328. https://doi.org/10.3390/plants10112328
Dabravolski SA, Isayenkov SV. New Insights into Plant TPK Ion Channel Evolution. Plants. 2021; 10(11):2328. https://doi.org/10.3390/plants10112328
Chicago/Turabian StyleDabravolski, Siarhei A., and Stanislav V. Isayenkov. 2021. "New Insights into Plant TPK Ion Channel Evolution" Plants 10, no. 11: 2328. https://doi.org/10.3390/plants10112328
APA StyleDabravolski, S. A., & Isayenkov, S. V. (2021). New Insights into Plant TPK Ion Channel Evolution. Plants, 10(11), 2328. https://doi.org/10.3390/plants10112328






