Abstract
Chickpea (Cicer arietinum L.) is a major pulse crop in Israel grown on about 3000 ha spread, from the Upper Galilee in the north to the North-Negev desert in the south. In the last few years, there has been a gradual increase in broomrape infestation in chickpea fields in all regions of Israel. Resistant chickpea cultivars would be simple and effective solution to control broomrape. Thus, to develop resistant cultivars we screened an ethyl methanesulfonate (EMS) mutant population of F01 variety (Kabuli type) for broomrape resistance. One of the mutant lines (CCD7M14) was found to be highly resistant to both Phelipanche aegyptiaca and Orobanche crenata. The resistance mechanism is based on the inability of the mutant to produce strigolactones (SLs)—stimulants of broomrape seed germination. LC/MS/MS analysis revealed the SLs orobanchol, orobanchyl acetate, and didehydroorobanchol in root exudates of the wild type, but no SLs could be detected in the root exudates of CCD7M14. Sequence analyses revealed a point mutation (G-to-A transition at nucleotide position 210) in the Carotenoid Cleavage Dioxygenase 7 (CCD7) gene that is responsible for the production of key enzymes in the biosynthesis of SLs. This nonsense mutation resulted in a CCD7 stop codon at position 70 of the protein. The influences of the CCD7M14 mutation on chickpea phenotype and chlorophyll, carotenoid, and anthocyanin content were characterized.
1. Introduction
Chickpea (Cicer arietinum L.) is an important legume crop grown on over 10 million ha in at least 37 countries worldwide, including India (65%), Pakistan (10%), Iran (8%), and Turkey (5.5%). [1]. In Israel chickpea is one of the main legume crops, grown on about 3000 ha with an average yield of about 3.5 t/ha. In recent years, the two broomrape species in Israel, Egyptian broomrape (Phelipanche aegyptiaca Pers.) and crenate broomrape (Orobanche crenata Forsk.), have become a major problem in chickpea field production [2]. The only broomrape-control methods that have been successfully utilized commercially in other crops are resistant varieties and chemical control [3,4,5].
Broomrapes (Phelipanche spp. and Orobanche spp.) are worldwide weedy root parasites of dicotyledonous crops, causing severe losses in the yield and quality of agricultural crops [6,7]. The initial step of broomrape–plant recognition involves root-exuded strigolactones (SLs), which have long been known to induce broomrape seed germination [8,9], and have been recently recognized as plant hormones affecting plant development and growth [10]. SLs consist of a tricyclic lactone (A, B, and C rings) connected to a butenolide group (D ring) via an enol ether bridge. SLs’ degree of activity, function, and specificity depend on the various substituents on the A and B rings [11]. The SL biosynthesis pathway in plants is derived from the carotenoid pathway [12,13,14], in which β-carotene is converted into carlactone by three catalytic enzymes: D-27 (9-cis/all-trans-β-carotene isomerase) [15], and two carotenoid cleavage dioxygenases, CCD7 and CCD8 [16,17]. Carlactone is converted to SLs by the cytochrome P450 monooxygenase- homolog activity of MORE AXILLARY GROWTH1 (MAX1) in rice [18], and MAX1 and lateral branching oxidoreductase in Arabidopsis [16,19,20]. SLs are produced mainly in roots and their active transport to the rhizosphere by the exporter pleiotropic drug resistance 1 (PDR1), identified in Petunia, was shown [21,22,23].
In a previous study, we obtained a tomato CCD7-deletion mutant showing broomrape resistance [24,25]. SL-deficient sorghum and rice mutants also demonstrate high degrees of resistance to Striga spp. [26,27]. Moreover, resistance to parasitic weeds based on low SL exudation exists in pea and faba bean germplasms [28,29]. Mutants defective in SL biosynthesis are characterized by a highly branched/tillering phenotype [30,31]. Furthermore, SLs regulate root architecture [16,32,33,34,35].
The objectives of the present study were to isolate and characterize an ethyl methanesulfonate (EMS)-mutagenized F01 chickpea mutant, CCD7M14, which shows considerable resistance to broomrape, and to elucidate its resistance mechanism, characterize its phenotype, and determine its leaf chlorophyll, carotenoid and anthocyanin contents.
2. Results
2.1. Mutagenesis and Screening for Broomrape Resistance
EMS mutagenesis was applied to seeds of a wild-type (WT) F01 chickpea breeding line (Kabuli type), and 3000 families of the second generation were tested for resistance to both P. aegyptiaca and O. crenata. A chickpea mutant showing high resistance to both broomrapes, was identified—CCD7M14 (Figure S1).
2.2. Phenotyping
2.2.1. Resistance to P. aegyptiaca and O. crenata
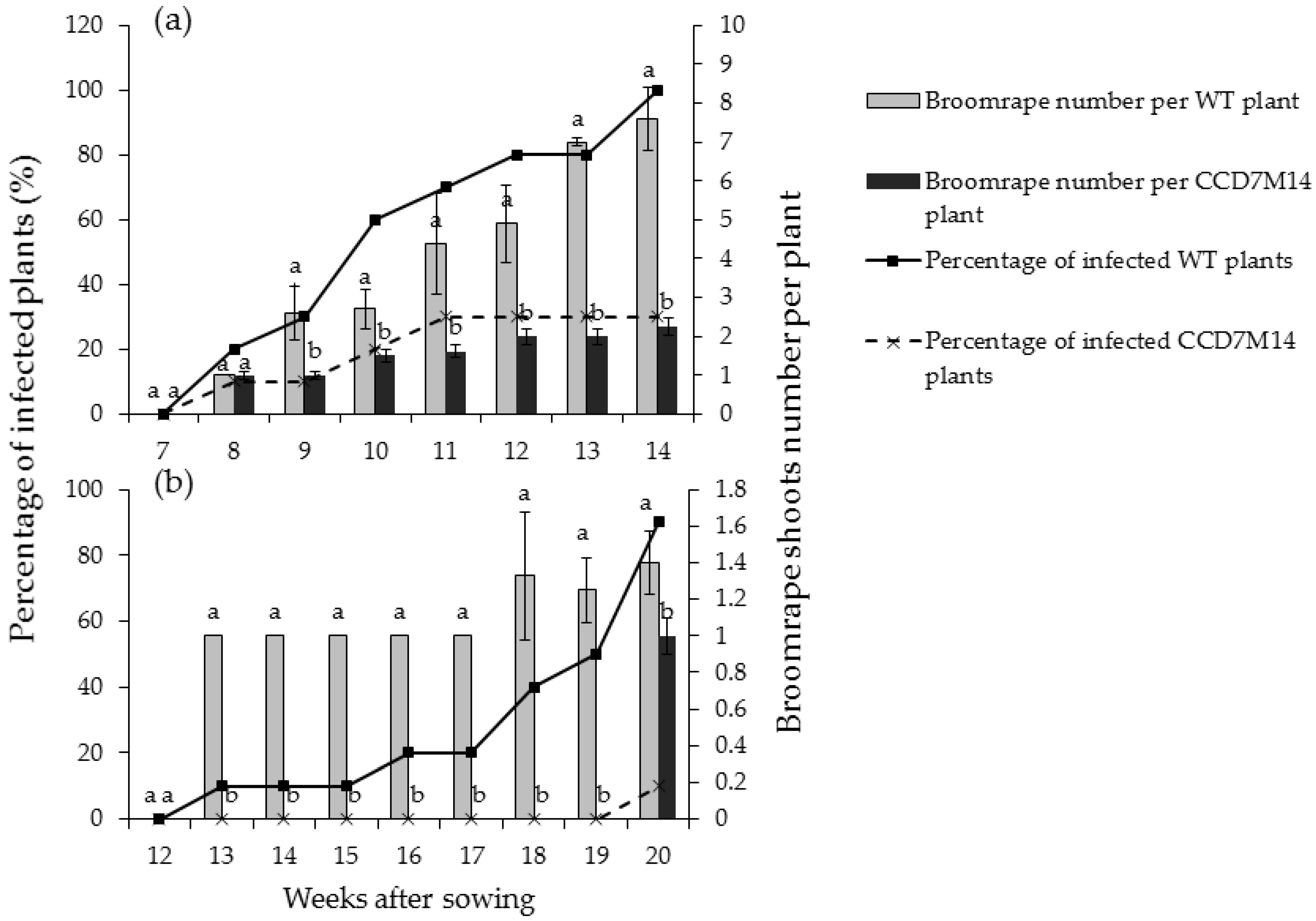
P. aegyptiaca shoots began to emerge aboveground 8 weeks after sowing in pots with WT F01 plants. At this time, about 20% of the WT F01 plants were infected with one or two shoots (Figure 1a). Both shoot number above the soil and percentage of infected plants increased rapidly over time, and at the end of the experiment (14 weeks), all WT F01 plants were infected with 8–10 aboveground shoots. At this time only one broomrape shoot was observed in two pots planted with CCD7M14 (percentage of infected plants was 20%). Throughout the course of the experiment, both percentage of infected plants and number of aboveground shoots per plant were significantly lower for the mutant plants. The roots were washed and broomrape number and biomass were recorded. About 16.10 ± 4.23 broomrape shoots were counted per WT F01 plant with average biomass of 82.11 ± 6.69 g, whereas only 1.60 ± 1.78 shoots with total biomass of 7.93 ± 5.15 g were found per mutant plant (Table 1).
Figure 1.
Aboveground broomrape shoots in pots planted with WT F01 or the CCD7M14 mutant. The experiments were arranged in a completely randomized design with 10 replications (pots) per treatment. Lines show the percentages of infected plants, bars indicate the average numbers of aboveground shoots attached to the infected plants. (a) Infestation with P. aegyptiaca. (b) Infestation with O. crenata. Vertical lines indicate standard error of the mean (SEM). Lowercase letters indicate least-significant differences (LSD), based on the Tukey–Kramer honestly significant difference test (α = 0.05) between the chickpea lines.

Table 1.
Statistical analysis of the chickpea resistance experiments. The results were subjected to ANOVA. The experiments were conducted with ten replications. SEM—standard error of the mean, dF—Degrees of Freedom; F—F ratio, Prob > F—F probability.
O. crenata developed more slowly than P. aegyptiaca. First O. crenata shoots emerged aboveground 12 weeks after planting in WT F01 pots (Figure 1b). At the end of the experiment (20 weeks after sowing), 90% of WT F01 plants were infected with one or two shoots. About 13.6 ± 3.48 broomrapes with a total biomass of about 109.74 ± 10.92 g per WT F01 plant were observed after root washing (Table 1). CCD7M14 plants were highly resistant to O. crenata. Only one aboveground shoot was observed in one pot at the end of the experiment, and about 2.20 ± 1.71 broomrapes with a total biomass of 13.78 ± 6.96 g were counted on the washed roots (Table 1).
2.2.2. Resistance Mechanism
To determine whether the resistance mechanism of CCD7M14 was based on its inability to synthesize SLs or secrete them into the rhizosphere, we tested its ability to stimulate broomrape seed germination. P. aegyptiaca seed germination near the WT F01 root system was high (76.84 ± 6.28%), whereas in the pots with CCD7M14 plants, only 0.72 ± 0.45% of the seeds germinated. Germination of O. crenata seeds was about 42.12 ± 2.57% in the pots with the WT, whereas in the mutant pots, no O. crenata seed germination was observed (Table 2).

Table 2.
Statistical analysis of the broomrape seed germination closed to chickpea roots. The results were subjected to ANOVA. The experiments were conducted with five replications. SEM—standard error of the mean, dF—Degrees of Freedom; F—F ratio, Prob > F—F probability.
The ability of WT and CCDM14 root exudates to stimulate P. aegyptiaca seed germination was tested in vitro in Petri dishes. Root exudate of the WT applied to the seeds at concentrations of 0.1, 1 and 10 μL/mL caused P. aegyptiaca germination at rates of 28.1 ± 5.78, 77.38 ± 3.13, and 84.84 ± 4.28%, respectively (compared to 79.19 ± 1.7% following application of 10−6 M GR24, a synthetic SL, as a positive control) (Table 3). A low percentage of seed germination was induced by the mutant root exudates (9.02 ± 0.77, 15.94 ± 1.19, and 34.95 ± 2.52% at concentrations of 0.1, 1 and 10 μL/mL, respectively), but only short radicals developed, which did not continue to elongate normally and were dead after 1 week.

Table 3.
Statistical analysis of the broomrape seed germination caused by root exudates. The results were subjected to ANOVA. The experiments were conducted with five replications. SEM—standard error of the mean, dF—Degrees of Freedom; F—F ratio, Prob > F—F probability.
Analysis of SLs in root exudates of WT F01 and CCD7M14 plants revealed the presence of orobanchol, orobanchyl acetate, and putative didehydroorobanchol in WT F01 root exudates, but no SLs in the mutant root exudates (Figure S2).
2.2.3. Plant Morphology and Pigment Contents
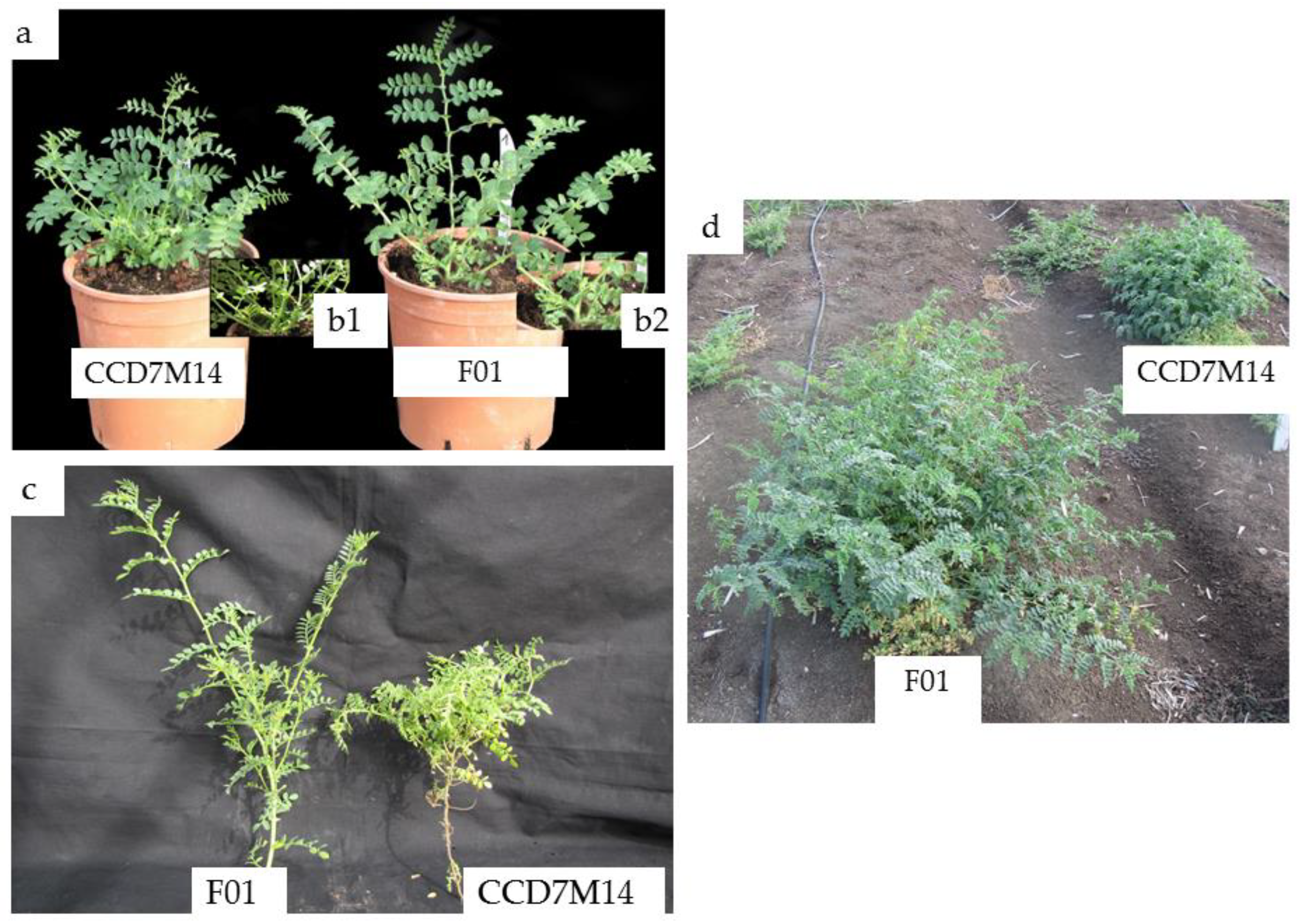
CCD7M14 plants had a SL-deficiency phenotype, with a high number of short branches compared to WT F01 plants. No significant differences in foliage or root biomass were found between the lines (Table 4). The CCD7M14 plants had 83% more primary branches than the WT F01 plants, and the mutant’s primary branch length was only 66% of that of the WT F01 plant. These morphological changes in CCD7M14 were observed both in the net house and under field conditions (Figure 2a–d), leading to a bushy shape at plant maturity.

Table 4.
Morphological characteristics of CCD7M14 compared to WT F01 chickpea.

Figure 2.
Morphological differences between WT F01 and CCD7M14. (a) One-month-old WT F01 (right) and CCD7M14 (left) plants grown in a net house. (b1,b2) Stem distribution on the lower section of the plants. (c) Primary branches of WT F01 (left) and CCD7M14 (right) plants. (d) Three-month-old WT F01 (left) and CCD7M14 (right) plants in the field.
Analysis of carotenoid, chlorophyll, and anthocyanin contents in the first, third, and fifth leaves revealed significant decreases in carotenoids and chlorophylls, and an increase in anthocyanins in the mutant as compared to its parental line (Table 5).

Table 5.
Contents of carotenoids, chlorophyll, and anthocyanins (μg per 1 g of fresh leaf biomass) in the leaves of WT F01 and CCD7M14.
2.3. DNA Analysis
Blast analyses of the chickpea genome based on the tomato CCD7 sequence revealed a single CCD7 gene with 64.9% protein sequence identity to the tomato protein (Figure 3). DNA sequence analysis of the CCD7 gene in CCD7M14 compared to the WT F01 line revealed a single G-to-A nucleotide transition at position 210 (Figure 4). This mutation led to stop-codon formation (*) instead of tryptophan (W) at amino acid position 70 (84 in tomato) (Figure 5). No other mutations were found in the chickpea CCD7 gene.

Figure 3.
Chickpea and tomato CCD7 protein sequence homology. The upper and lower sequences are of chickpea and tomato CCD7, respectively. Identical amino acids are indicated by a solid line, and similar amino acids are indicated by one or two dots according to their similarity levels. The mutated amino acid in CCD7 of broomrape-resistant line CCD7M14, at position 70 (84 according to tomato) is indicated in yellow and marked by an asterisk.

Figure 4.
The Blast results of DNA sequences (nucleotides 202–246) of WT F01 chickpea (upper line) and CCD7M14 (lower line). The G-to-A transition at position 210 is indicated in bold red letters.
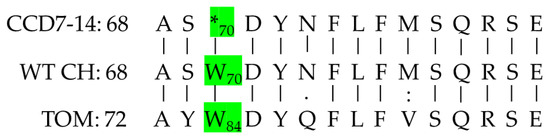
Figure 5.
The Blast results of the protein sequences (amino acids 68–82 (82–96 in tomato)) of CCD7M14 (CCD7-14), F01 (WT CH), and tomato (TOM). The W-to-stop codon (*) transition at position 70 (84 in tomato) is indicated in green.
3. Discussion
Chickpea mutant CCD7M14 was produced by EMS mutagenesis. The mutant showed high resistance to both P. aegyptiaca and O. crenata (Figure S1). Only one mutant plant was infected with a single P. aegyptiaca, and one with a single O. crenata shoots in all experiments, compared to 90–100% infection in WT F01 plants with more than 8–10 aboveground broomrape shoots (Figure 1a,b). However, once an attachment formed on the mutant roots, parasite development progressed normally. Since no P. aegyptiaca or O. crenata seed germination was observed near CCD7M14 roots, and its root exudates did not stimulate their seed germination in Petri dishes, it is suggested that the CCD7M14 resistance mechanism is based on its inability to synthesize SLs or to secrete them into the rhizosphere. Indeed, DNA sequence analysis of the CCD7M14 CCD7 gene revealed stop-codon formation due to a single G-to-A nucleotide transition at position 210 (Figure 4 and Figure 5). This resulted in the absence of the SLs orobanchol, orobanchyl acetate, and didehydroorobanchol in the root exudates (Figure S2), rendering the mutant plant resistant to the parasite because no seed germination could occur near its roots. This resistance mechanism has been reported in tomato [25,36,37,38], pea [39] and faba bean [30,40]. Previously, this type of resistance had been obtained by fast-neutron mutagenesis [24,25] and targeted mutagenesis [37,38]. It had also been found in wild tomato species (Solanum pennellii [36]), and recognized in resistant cultivars of faba bean and pea [30,39,40]. In our case, the resistance was obtained by EMS mutagenesis, where one point mutation in the CCD7 gene resulted in the formation of a stop codon, leading to the same results as CCD7 deletion by fast-neutron mutagenesis [25,26] or silencing of CCD8 using CRISPR/Cas9-mediated mutagenesis [37,38]. It is important to note that to date, all identified CCD7 genes have been single copies, in contrast to two, four and six copies of CCD8 identified in maize, rice and sorghum, respectively [41].
It has been shown that plants exude mixtures of several SLs, and every plant species is characterized by a specific SL profile [42]. In the current study, we first identified the SLs produced by chickpea roots. LC/MS/MS analysis revealed that the WT F01 chickpea cultivar produces three SLs: orobanchol, orobanchyl acetate, and putative didehydroorobanchol isomer(s). All three belong to the orobanchol type, which only differs from the strigol-type SLs in the stereochemistry of the C-ring [43], and are derived from 4-deoxyorobanchol in rice [18]. Some other species, such as Populus, pea, petunia, and tomato, have been reported to have only orobanchol-type SLs [44]. Orobanchol, first isolated from red clover (Trifolium pratense L.) root exudates [9], is probably the most abundant hydroxy-SL in the plant kingdom [42]. This SL assumes to be a central intermediate in SL biosynthesis, and it has been suggested as a precursor of other SL molecules, such as: fabacol, orobanchyl acetate, solanacol, and so on [43]. Putative didehydroorobanchol has been detected in root exudates of tomato [26], tobacco [42], and Medicago truncatula [45]; and orobanchyl acetate in red clover [46], rice and tobacco [47]. The didehydroorobanchol isomer in M. truncatula was named medicaol [45]. Both orobanchol and orobanchyl acetate have been reported to be produced by Asteraceae plants and by faba bean [48,49].
CCD7M14 was characterized by a typical SL-deficient phenotype—increased branching and reduced plant height. These results are in agreement with Vogel et al. [50], where transgenic tomato plants expressing their endogenous CCD7 gene in the antisense form also displayed increased branching and reduced plant height. Similar observations have also been reported for pea [51], petunia [52], poplar [53], and Arabidopsis [54]. According to Boyer et al. [11], orobanchyl acetate and 5- deoxystrigol are more active at inhibiting shoot branching than strigol and orobanchol. Furthermore, blockage of orobanchol biosynthesis from carlactonoic acid in tomato did not rescue the branching phenotype [55]. The absence of orobanchyl acetate in CCD7M14 plants likely explains its bushy shape at maturity (Figure 5). Orobanchol and putative didehydroorobanchol may be involved in other biological processes, such as regulation of photosynthesis and pigment accumulation. We found significant decreases in chlorophyll and carotenoid contents and an increase in anthocyanins in the leaves of CCD7M14 as compared to the WT F01 line (Table 2). Exogenous application of the synthetic SL GR24 under stress conditions has been shown to control chlorophyll degradation and maintain the photosynthetic rate [56,57,58,59]. On the other hand, chlorophyll content in sunflower leaves was not influenced by GR24 treatment of achene pre-sowing, but carotenoid content increased [60]. GR24 has been found to affect ABA-induced activation of anthocyanin biosynthesis in grapevine berries [61]. In transgenic tobacco lines impaired in SL biosynthesis, overaccumulation of anthocyanins in the mature stems likely results from antagonism between the SL and jasmonic acid pathways [62]. SL regulation of anthocyanin accumulation has been shown in Arabidopsis [63,64].
4. Materials and Methods
4.1. Plant Material
All experiments were carried out with: (a) a WT F01 chickpea breeding line (Kabuli type) that is erect, produces high yields, and is resistant to both Fusarium wilt and Ascochyta blight and (b) CCD7M14, a chickpea EMS mutant line derived from WT F01. Broomrape seeds were collected from P. aegyptiaca and O. crenata inflorescences parasitizing tomato plants grown in Kibbutz Bet Ha’shita (32°33′15″ N 35°26′15″ E) and chickpea plants grown in Kibbutz Kfar H’horesh (32°42′7.56″ N 35°16′27.47″ E), respectively. The inflorescences were dried at 23–35 °C for 2 months and then the seeds were separated with a 300-mesh size sieve (50 µm) and stored in the dark at 4 °C until use.
4.2. Mutagenesis
WT F01 chickpea breeding line seeds were used for mutagenesis. Approximately 6000 WT F01 seeds were allowed to swell in water for 10 h and then exposed to the mutation inducer EMS at a concentration of 4% (vol/vol) which, according to the dose-response curve, decreased seed germination by 50%. After shaking at 50 rpm for 10 h, the EMS was removed, and the seeds were washed under running tap water for 14 h. The seeds were dried under airflow for 48 h and delivered to Shorashim Nursery Ltd., Israel, to produce seedlings. The seedlings were planted and grown in a field at the Western Galilee experimental farm, Israel (32°55′ N 35°04′ E), to produce M2 seeds.
4.3. Screening for Broomrape Resistance
An EMS-mutated population of about 3000 families (each derived from a single M1 plant) was used to screen for broomrape resistance. Eight M2 generation seeds from each family were seeded separately in soil containing seeds of P. aegyptiaca and O. crenata at a concentration of 20 mg seeds per kg of soil (~3000 seeds/kg). After 3 months, plant roots were evaluated for broomrape infection. Families of plants that were free of broomrape were selected for the next screening, leading to identification of the broomrape-resistant mutant CCD7M14.
4.4. Phenotype Determination
4.4.1. Evaluation of Broomrape Resistance
Broomrape-resistance tests were conducted in 2 L pots, each filled with soil mixed with the seeds of P. aegyptiaca and O. crenata at a concentration of 20 mg seeds per kg soil. Control pots did not contain broomrape seeds. Each pot was planted with one chickpea plant. Organic medium-heavy clay–loam soil collected in Newe Ya’ar Research Center (32°42′9″ N, 35°10′9″ E) was used in all experiments. The plants were grown in nethouse and irrigated and fertilized as needed. The experiments were arranged in a completely randomized design with 10 replications (pots) per treatment. Once a week, the number of broomrape shoots per pot was evaluated. At the end of the experiments, the roots were gently washed out of the pots under tap water and broomrape number and fresh biomass were determined.
4.4.2. Resistance Mechanism Determination
The ability of WT and mutant plants to induce germination of P. aegyptiaca and O. crenata seeds was tested in GF/A glass microfiber filter paper envelopes [25]. Briefly, P. aegyptiaca or O. crenata seeds inside the paper envelopes were placed close to the chickpea roots at planting. Seed germination percentage was recorded four weeks after planting using a stereoscopic microscope. Control pots (without plants) were used for spontaneous seed germination determination.
To analyze SLs in root exudates, WT F01 and CCD7M14 plants were grown under hydroponic conditions with feeding solution circulated through activated charcoal [25]. Once a week, the charcoal was washed with water and extracted with acetone. The acetone solutions were combined and evaporated under reduced pressure at 35 °C (Rotavapor, Büchi, Switzerland) from all samples. The residue was dissolved in 200 mL water and the solution was extracted three times with equal volumes of ethyl acetate. The ethyl acetate fractions were combined, washed with 0.2 M K2HPO4 (pH 8.3), dried over anhydrous Na2SO4, and concentrated under reduced pressure at 35 °C. Dry extracts were stored at 4 °C.
Samples of root exudates were tested for the ability to germinate preconditioned P. aegyptiaca seeds according to Yoneyama et al. (2007) [65]. Briefly, dried root exudates were dissolved in methanol up to concentration of 0.2, 2 and 20 μg/mL of which 100 μL was applied to filter paper inside 45-mm diameter Petri dishes. After drying under air flour, 0.2 mL of sterile water was added to the disks to get final concentrations of 0.1, 1, and 10 μg/mL. Disinfected P. aegyptiaca seeds were distributed on a 45 mm filter paper disk and kept moistened for 1 week. Then the disks with seeds on them were dried gently on sterile filter paper and transferred to the Petri dishes upon the disks containing root exudates. For the positive control, stimulation with GR24 at a concentration of 10−6 M was used. The plates were kept at 25 °C for 10 days, and the P. aegyptiaca seed germination was evaluated utilizing of a stereoscopic microscope.
LC-MS/MS analysis of proton adduct ions of SLs was performed with a triple quadrupole/linear ion trap instrument (LIT) (QTRAP5500; AB Sciex) with an electrospray source according to Yoneyama et al., 2007 [65]. All peaks corresponding to strigolactones were confirmed by P. aegyptiaca seed-germination assay [25].
4.4.3. Plant Morphology and Pigment Contents
The plants of WT F01 and CCD7M14 were grown in 4 L pots in Newe Ya’ar organic soil. After 14 weeks, the plants were harvested by cutting the stems at the pot’s soil surface. First, third and fifth leaves were sampled for determination of total carotenoid, anthocyanin, and chlorophyll a and b contents. The number of primary branches, the number of secondary branches per primary branch, and foliage and root fresh biomass were determined.
Contents of carotenoids and anthocyanin were measured according to Segev et al. [66], and chlorophyll were was measured according to Lichtenthaler [67]. Briefly, chlorophyll and anthocyanin were extracted using methanol and acidic methanol (99% methanol and 1% hydrochloric acid), respectively. The test tubes were incubated at room temperature for two days in the dark. After two days, the solutions were tested in a spectrophotometer at 665, 652, 530, and 470 nm wavelengths. From the data we calculated the relative amounts of total chlorophyll, chlorophyll a, chlorophyll b, total carotenoids, and total anthocyanins according to the following formulas:
Chlorophyll A (μg/mL) = 16.72 × A665 − 9.16 × A652;
Chlorophyll B (μg/mL) = 34.09 × A652 − 15.28 × A665;
Total chlorophyll (a + b) (μg/mL) = 1.44 × A665 + 24.93 × A652;
Total carotenoids (μg/mL) = (1000 × A470 − 1.63 × Chlorophyll A − 104.96 × Chlorophyll B)/221;
Total anthocyanins (μg/mL) = (449.1 × A530 + 24.93 × 2000)/24,500.
Chlorophyll B (μg/mL) = 34.09 × A652 − 15.28 × A665;
Total chlorophyll (a + b) (μg/mL) = 1.44 × A665 + 24.93 × A652;
Total carotenoids (μg/mL) = (1000 × A470 − 1.63 × Chlorophyll A − 104.96 × Chlorophyll B)/221;
Total anthocyanins (μg/mL) = (449.1 × A530 + 24.93 × 2000)/24,500.
The final results were calculated in μg per 1 g of fresh leaf biomass.
4.5. DNA Extraction and PCR Amplification
Total genomic DNA was extracted from young leaves of 2-week-old M3 plants homozygous for broomrape resistance. Primer design, PCR amplification, electrophoresis in a 1.0% agarose gel, and sequence analysis of the CCD7 gene were performed as described by Schreiber et al. [68], with several modifications: annealing was performed at 55 °C for 30 s and synthesis at 72 °C for 60 s. Eight pairs of primers, purchased from Syntezza Bioscience Ltd. (Jerusalem, Israel), were used (Table 6).

Table 6.
Primer sets used in this study.
4.6. Statistical Analysis
All experimental results were subjected to ANOVA utilizing JMP software, version 5.0 (SAS Institute Inc., Cary, NC, USA). Data on seed germination were separated by standard error of the mean (SEM). To meet the assumption of ANOVA, percentage data were arcsine-transformed before analysis. The results on the number of aboveground broomrape shoots were compared by SEM and by least-significant differences (LSD), based on Tukey–Kramer honestly significant difference test (α = 0.05). Data on the number and biomass of P. aegyptiaca and O. crenata attached to chickpea roots after root washing were separated by SEM. The experiments on chickpea lines sensitivity to P. aegyptiaca and O. crenata were repeated twice. The repeated experiments were compared using Fisher’s t-test, which showed homogeneity of variances; therefore, the data were combined. The test of the differences in morphology between WT F01 and CCD7M14 and the data of pigment concentration in chickpea leaves was conducted with 5 and 3 replicates, respectively and separated by SEM. The results were analyzed by LS means contrast test (α = 0.05).
5. Conclusions
Using EMS mutagenesis, chickpea line CCD7M14 showing high resistance to both O. crenata and P. aegyptiaca was developed. The resistance mechanism was based on blockage of SL synthesis, probably caused by stop codon formation due to the point mutation in the CCD7 gene. Root exudates of the mutant did not contain SLs. The mutant plants displayed increased branching and reduced plant height; decreased chlorophyll and carotenoid contents; and increased accumulation of anthocyanin in the leaves compared with the WT.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10122552/s1, Figure S1: WT F01 (right) and CCD7M14 (left) plants growing in soil mixed with seeds of P. aegyptiaca at a concentration of 20 mg seeds per kg soil. Figure S2: Selected reaction monitoring (SRM) chromatograms of WT F01 (a) and CCD7M14 (b) root exudates. Determination of SLs was based on the retention time and transition of m/z 345 > 97 for didehydroorobanchol and 347 > 233 for orobanchol and orobanchyl acetate.
Author Contributions
Conceptualization, S.G., J.H. and E.D.; Methodology, J.H. and E.D.; Software, S.G.; Investigation, E.S., O.A.-S., K.Y., X.X. and A.B.; Resources, S.G. and J.H.; Data Curation, E.D.; Writing—Original Draft Preparation, S.G., J.H. and E.D.; Writing—Review and Editing, S.G., J.H., K.Y. and E.D.; Visualization, S.G. and E.S.; Supervision, S.G. and E.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Datais contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAOSTAT. Available online: http://faostat.fao.org/default.aspx (accessed on 1 January 2016).
- Galili, S.; Hovav, R.; Dor, E.; Hershenhorn, J.; Harel, A.; Amir-Segev, O.; Bellalou, A.; Badani, H.; Smirnov, E.; Achdari, G. The history of chickpea cultivation and breeding in Israel. Isr. J. Plant Sci. 2018, 65, 186–194. [Google Scholar] [CrossRef]
- Dor, E.; Smirnov, E.; Galili, S.; Guy, A.; Hershenhorn, J. Characterization of the novel tomato mutant HRT, resistant to acetolactate synthase–inhibiting herbicides. Weed Sci. 2016, 64, 348–360. [Google Scholar] [CrossRef]
- Dor, E.; Galili, S.; Smirnov, E.; Hacham, Y.; Amir, R.; Hershenhorn, J. The effects of herbicides targeting aromatic and branched chain amino acid biosynthesis support the presence of functional pathways in broomrape. Front. Plant Sci. 2017, 8, 707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venezian, A.; Dor, E.; Achdari, G.; Plakhine, D.; Smirnov, E.; Hershenhorn, J. The influence of the plant growth regulator maleic hydrazide on Egyptian broomrape early developmental stages and its control efficacy in tomato under greenhouse and field conditions. Front. Plant Sci. 2017, 8, 691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, C.; Riches, C.R. Parasitic Weeds of the World: Biology and Control; CAB International: Wallingford, UK, 1993. [Google Scholar]
- Joel, D.M.; Hershenhorn, J.; Eizenberg, H.; Aly, R.; Ejeta, G.; Rich, P.J.; Ransom, J.K.; Sauerborn, J.; Rubiales, D. Biology and management of weedy root parasites. In Horticultural Reviews; John Wiley & Sons: Hoboken, NJ, USA, 2007; Volume 33. [Google Scholar] [CrossRef]
- Joel, D.M.; Kleifeld, Y.; Losner-Goshen, D.; Herzlinger, G.; Gressel, J. Transgenic crops against parasites. Nature 1995, 374, 220–221. [Google Scholar] [CrossRef]
- Yokota, T.; Sakai, H.; Okuno, K.; Yoneyama, K.; Takeuchi, Y. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 1998, 49, 1967–1973. [Google Scholar] [CrossRef]
- Xie, X.; Yoneyama, K.; Yoneyama, K. The strigolactone story. Annu. Rev. Phytopathol. 2010, 48, 93–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, F.-D.; de Saint Germain, A.; Pillot, J.-P.; Pouvreau, J.-B.; Chen, V.X.; Ramos, S.; Stévenin, A.; Simier, P.; Delavault, P.; Beau, J.-M.; et al. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: Molecule design for shoot branching. Plant Physiol. 2012, 159, 1524–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorefan, K.; Booker, J.; Haurogné, K.; Goussot, M.; Bainbridge, K.; Foo, E.; Chatfield, S.; Ward, S.; Beveridge, C.; Rameau, C.; et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matusova, R.; Rani, K.; Verstappen, F.W.A.; Franssen, M.C.R.; Beale, M.H.; Bouwmeester, H.J. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005, 139, 920–934. [Google Scholar] [CrossRef] [Green Version]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.J.; Beyer, P.; Al-Babili, S. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. Lateral branching oxidorreductase acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Van Dijk, A.D.J.; Scaffidi, A.; Flematti, G.R.; Hofmann, M.; Charnikhova, T.; Verstappen, F.W.A.; Hepworth, J.; Van Der Krol, S.; Leyser, O.; et al. Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 2014, 10, 1028–1033. [Google Scholar] [CrossRef]
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089. [Google Scholar] [CrossRef] [Green Version]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [Green Version]
- Sasse, J.; Simon, S.; Gübeli, C.; Liu, G.-W.; Cheng, X.; Friml, J.; Bouwmeester, H.J.; Martinoia, E.; Borghi, L. Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr. Biol. 2015, 25, 647–655. [Google Scholar] [CrossRef] [Green Version]
- Delavault, P.; Montiel, G.; Brun, G.; Pouvreau, J.-B.; Thoiron, S.; Simier, P. Communication between host plants and parasitic plants. Adv. Bot. Res. 2017, 82, 55–82. [Google Scholar]
- Dor, E.; Alperin, B.; Wininger, S.; Ben-Dor, B.; Somvanshi, V.S.; Koltai, H.; Kapulnik, Y.; Hershenhorn, J. Characterization of a novel tomato mutant resistant to Orobanche and Phelipanche spp. weedy parasites. Euphytica 2010, 171, 371–380. [Google Scholar] [CrossRef]
- Dor, E.; Yoneyama, K.; Wininger, S.; Kapulnik, Y.; Yoneyama, K.; Koltai, H.; Xie, X.; Hershenhorn, J. Strigolactone deficiency confers resistance in tomato line SL-ORT1 to the parasitic weeds Phelipanche and Orobanche spp. Phytopathology 2011, 101, 213–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ejeta, G. Breeding for Striga resistance in sorghum: Exploitation of an intricate host–parasite biology. Crop Sci. 2007, 47, S216–S227. [Google Scholar] [CrossRef]
- Jamil, M.; Rodenburg, J.; Charnikhova, T.; Bouwmeester, H.J. Pre-attachment Striga hermonthica resistance of new rice for Africa (NERICA) cultivars based on low strigolactone production. New Phytol. 2011, 192, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Fondevilla, S.; Fernández-Aparicio, M.; Satovic, Z.; Emeran, A.A.; Torres, A.M.; Moreno, M.T.; Rubiales, D. Identification of quantitative trait loci for specific mechanisms of resistance to Orobanche crenata Forsk. in pea (Pisum sativum L.). Mol. Breed. 2010, 25, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Aparicio, M.; Kisugi, T.; Xie, X.; Rubiales, D.; Yoneyama, K. Low strigolactone root exudation: A novel mechanism of broomrape (Orobanche and Phelipanche spp.) resistance available for faba bean breeding. J. Agric. Food Chem. 2014, 62, 7063–7071. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef] [Green Version]
- Kapulnik, Y.; Delaux, P.-M.; Resnick, N.; Mayzlish-Gati, E.; Wininger, S.; Bhattacharya, C.; Séjalon-Delmas, N.; Combier, J.-P.; Bécard, G.; Belausov, E.; et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 2011, 233, 209–216. [Google Scholar] [CrossRef]
- Koltai, H.; Dor, E.; Hershenhorn, J.; Joel, D.M.; Weininger, S.; Lekalla, S.; Shealtiel, H.; Bhattacharya, C.; Eliahu, E.; Resnick, N.; et al. Strigolactones’ effect on root growth and root-hair elongation may be mediated by auxin-efflux carriers. J. Plant Growth Regul. 2010, 29, 129–136. [Google Scholar] [CrossRef]
- Koltai, H.; LekKala, S.P.; Bhattacharya, C.; Mayzlish-Gati, E.; Resnick, N.; Wininger, S.; Dor, E.; Yoneyama, K.; Yoneyama, K.; Hershenhorn, J.; et al. A tomato strigolactone-impaired mutant displays aberrant shoot morphology and plant interactions. J. Exp. Bot. 2010, 61, 1739–1749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Wei, Q.; Shu, J.; Gan, Z.; Li, B.; Yan, D.; Huang, Z.; Guo, Y.; Wang, X.; Zhang, L. Exploration of resistance to Phelipanche aegyptiaca in tomato. Pest Manag. Sci. 2020, 76, 3806–3821. [Google Scholar] [CrossRef]
- Bari, V.K.; Nassar, J.A.; Kheredin, S.M.; Gal-On, A.; Ron, M.; Britt, A.; Steele, D.; Yoder, J.; Aly, R. CRISPR/Cas9-mediated mutagenesis of Carotenoid Cleavage Dioxygenase 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019, 9, 11438. [Google Scholar] [CrossRef] [PubMed]
- Bari, V.K.; Nassar, J.A.; Meir, A.; Aly, R. Targeted mutagenesis of two homologous ATP-binding cassette subfamily G (ABCG) genes in tomato confers resistance to parasitic weed Phelipanche aegyptiaca. J. Plant Res. 2021, 134, 585–597. [Google Scholar] [CrossRef]
- Pavan, S.; Schiavulli, A.; Marcotrigiano, A.R.; Bardaro, N.; Bracuto, V.; Ricciardi, F.; Charnikhova, T.; Lotti, C.; Bouwmeester, H.J.; Ricciardi, L. Characterization of low-strigolactone germplasm in pea (Pisum sativum L.) resistant to crenate broomrape (Orobanche crenata Forsk.). Mol. Plant-Microbe Interact. 2016, 29, 743–749. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Aparicio, M.; Moral, A.; Kharrat, M.; Rubiales, D. Resistance against broomrapes (Orobanche and Phelipanche spp.) in faba bean (Vicia faba) based in low induction of broomrape seed germination. Euphytica 2012, 186, 897–905. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Bradbury, L.M.T.; Wurtzel, E.T. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 2010, 504, 104–111. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, K.; Xie, X.; Yoneyama, K.; Takeuchi, Y. Strigolactones: Structures and biological activities. Pest Manag. Sci. 2009, 65, 467–470. [Google Scholar] [CrossRef]
- Wang, Y.; Bouwmeester, H.J. Structural diversity in the strigolactones. J. Exp. Bot. 2018, 69, 2219–2230. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Kusumoto, D.; Takeuchi, Y.; Yoneyama, K.; Yamada, Y.; Yoneyama, K. 2′-Epi-orobanchol and solanacol, two unique strigolactones, germination stimulants for root parasitic weeds, produced by tobacco. J. Agric. Food Chem. 2007, 55, 8067–8072. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, T.; Hayashi, H.; Akiyama, K. Medicaol, a strigolactone identified as a putative didehydro-orobancholisomer, from Medicago truncatula. Phytochemistry 2015, 111, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yoneyama, K.; Kusumoto, D.; Yamada, Y.; Yokota, T.; Takeuchi, Y.; Yoneyama, K. Isolation and identification of alectrol as (+)-orobanchyl acetate, a germination stimulant for root parasitic plants. Phytochemistry 2008, 69, 427–431. [Google Scholar] [CrossRef]
- Xie, X.; Yoneyama, K.; Kisugi, T.; Uchida, K.; Ito, S.; Akiyama, K.; Hayashi, H.; Yokota, T.; Nomura, T.; Yoneyama, K. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 2013, 6, 153–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneyama, K.; Xie, X.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. Characterization of strigolactones exuded by Asteraceae plants. Plant Growth Regul. 2011, 65, 495–504. [Google Scholar] [CrossRef] [Green Version]
- Trabelsi, I.; Yoneyama, K.; Abbes, Z.; Amri, M.; Xie, X.; Kisugi, T.; Kim, H.I.; Kharrat, M.; Yoneyama, K. Characterization of strigolactones produced by Orobanche foetida and Orobanche crenata resistant faba bean (Vicia faba L.) genotypes and effects of phosphorous, nitrogen, and potassium deficiencies on strigolactone production. South Afr. J. Bot. 2017, 108, 15–22. [Google Scholar] [CrossRef]
- Vogel, J.T.; Walter, M.H.; Giavalisco, P.; Lytovchenko, A.; Kohlen, W.; Charnikhova, T.; Simkin, A.J.; Goulet, C.; Strack, D.; Bouwmeester, H.J.; et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010, 61, 300–311. [Google Scholar] [CrossRef]
- De Saint Germain, A.; Ligerot, Y.; Dun, E.A.; Pillot, J.-P.; Ross, J.J.; Beveridge, C.A.; Rameau, C. Strigolactones stimulate internode elongation independently of gibberellins. Plant Physiol. 2013, 163, 1012–1025. [Google Scholar] [CrossRef] [Green Version]
- Snowden, K.C.; Simkin, A.J.; Janssen, B.J.; Templeton, K.R.; Loucas, H.M.; Simons, J.L.; Karunairetnam, S.; Gleave, A.P.; Clark, D.G.; Klee, H.J. The decreased apical dominance1/Petunia hybrida Carotenoid Cleavage Dioxygenase 8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 2005, 17, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Muhr, M.; Prüfer, N.; Paulat, M.; Teichmann, T. Knockdown of strigolactone biosynthesis genes in Populus affects branched 1 expression and shoot architecture. New Phytol. 2016, 212, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Ongaro, V.; Leyser, O. Hormonal control of shoot branching. J. Exp. Bot. 2008, 59, 67–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakabayashi, T.; Hamana, M.; Mori, A.; Akiyama, R.; Ueno, K.; Osakabe, K.; Osakabe, Y.; Suzuki, H.; Takikawa, H.; Mizutani, M.; et al. Direct conversion of carlactonoic acid to orobanchol by cytochrome P450 CYP722C in strigolactone biosynthesis. Sci. Adv. 2019, 5, eaax9067. [Google Scholar] [CrossRef] [Green Version]
- Min, Z.; Li, R.; Chen, L.; Zhang, Y.; Li, Z.; Liu, M.; Ju, Y.; Fang, Y. Alleviation of drought stress in grapevine by foliar-applied strigolactones. Plant Physiol. Biochem. 2019, 135, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ding, F.; Li, M.; Zhang, X.; Zhang, S.; Huang, B. Strigolactone and ethylene inhibitor suppressing dark-induced leaf senescence in perennial ryegrass involving transcriptional downregulation of chlorophyll degradation. J. Am. Soc. Hortic. Sci. 2021, 146, 79–86. [Google Scholar] [CrossRef]
- Qiu, C.-W.; Zhang, C.; Wang, N.-H.; Mao, W.; Wu, F. Strigolactone GR24 improves cadmium tolerance by regulating cadmium uptake, nitric oxide signaling and antioxidant metabolism in barley (Hordeum vulgare L.). Environ. Pollut. 2021, 273, 116486. [Google Scholar] [CrossRef]
- Zheng, X.; Li, Y.; Xi, X.; Ma, C.; Sun, Z.; Yang, X.; Li, X.; Tian, Y.; Wang, C. Exogenous strigolactones alleviate KCl stress by regulating photosynthesis, ROS migration and ion transport in Malus hupehensis Rehd. Plant Physiol. Biochem. 2021, 159, 113–122. [Google Scholar] [CrossRef]
- Sarwar, Y.; Shahbaz, M. Modulation in growth, photosynthetic pigments, gas exchange attributes and inorganic ions in sunflower (Helianthus annuus L.) by strigolactones (GR24) achene priming under saline conditions. Pak. J. Bot. 2020, 52, 23–31. [Google Scholar] [CrossRef]
- Ferrero, M.; Pagliarani, C.; Novák, O.; Ferrandino, A.; Cardinale, F.; Visentin, I.; Schubert, A. Exogenous strigolactone interacts with abscisic acid-mediated accumulation of anthocyanins in grapevine berries. J. Exp. Bot. 2018, 69, 2391–2401. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Joo, Y.; Cao, D.; Li, R.; Lee, G.; Halitschke, R.; Baldwin, G.; Baldwin, I.T.; Wang, M. Strigolactone signaling regulates specialized metabolism in tobacco stems and interactions with stem-feeding herbivores. PLoS Biol. 2020, 18, e3000830. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.; Yin, X.; Li, K.; Yang, Y.; Guo, J.; Miao, Y.; et al. Negative roles of strigolactone-related SMXL6, 7 and 8 proteins in drought resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef]
- Yoneyama, K.; Yoneyama, K.; Takeuchi, Y.; Sekimoto, H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 2007, 225, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Segev, A.; Badani, H.; Kapulnik, Y.; Shomer, I.; Oren-Shamir, M.; Galili, S. Determination of polyphenols, flavonoids, and antioxidant capacity in colored chickpea (Cicer arietinum L.). J. Food Sci. 2010, 75, S115–S119. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Schreiber, G.; Reuveni, M.; Evenor, D.; Oren-Shamir, M.; Ovadia, R.; Sapir-Mir, M.; Bootbool-Man, A.; Nahon, S.; Shlomo, H.; Chen, L.; et al. Anthocyanin1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the Anthocyanin Fruit phenotype of tomato. TAG 2012, 124, 295–307. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).