Genome-Wide Identification of the Thaumatin-like Protein Family Genes in Gossypium barbadense and Analysis of Their Responses to Verticillium dahliae Infection
Abstract
:1. Introduction
2. Results
2.1. Identification of TLP Genes in the Sea Island Cotton Genome
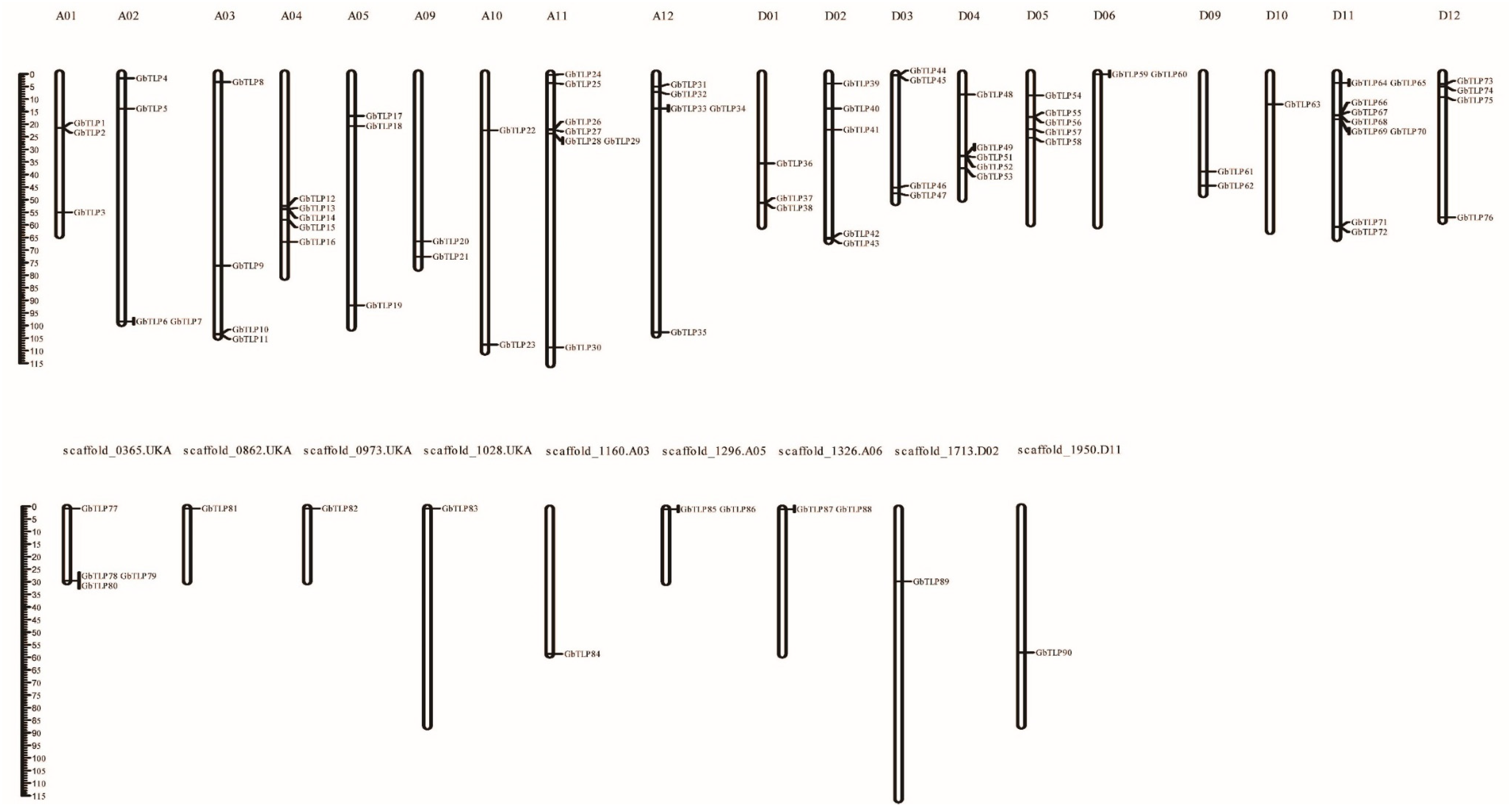
2.2. Chromosomal Distribution of GbTLPs and Gene Collinearity Analysis
2.3. Phylogenetic and Evolutionary Analysis
2.4. Motif Composition and Gene Structure Analyses
2.5. cis-Element Analysis of GbTLP Promoters
2.6. Identification of GbTLPs Targeted by miRNAs
2.7. Differential Expression of GbTLP Genes in Response to V. dahliae Infection
3. Discussion
3.1. Expansion of the GbTLP Gene Family in Sea Island Cotton
3.2. The Putative Regulatory Mechanisms of GbTLP Gene Expression
3.3. Expression Patterns of GbTLPs Responding to Verticillium Wilt Treatment
4. Materials and Methods
4.1. Identification and Annotation of Sea Island Cotton TLP Genes
4.2. Chromosomal Location Analysis
4.3. Synteny and Collinearity Analysis
4.4. Phylogenetic and Evolutionary Analysis
4.5. Gene Structure and Conserved Motif Analysis of the TLP Gene Family
4.6. Prediction of Regulatory cis-Elements in the Promoter of GbTLPs
4.7. Plant Materials and Treatments
4.8. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PR | Pathogenesis-related |
| TLP | Thaumatin-like protein |
| VD | Verticillium dahlia |
| pI | isoelectric point |
References
- Zhao, J.P.; Su, X.H. Patterns of molecular evolution and predicted function in thaumatin-like proteins of Populus trichocarpa. Planta 2010, 232, 949–962. [Google Scholar] [CrossRef]
- Liu, J.-J.; Sturrock, R.; Ekramoddoullah, A.K.M. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Reddy, A. Cloning and expression of a PR5-like protein from Arabidopsis: Inhibition of fungal growth by bacterially expressed protein. Plant Mol. Biol. 1997, 34, 949–959. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakrabarti, C. Crystal structure analysis of NP24-I: A thaumatin-like protein. Planta 2008, 228, 883–890. [Google Scholar] [CrossRef]
- Smole, U.; Bublin, M.; Radauer, C.; Ebner, C.; Breiteneder, H. Mal d 2, the thaumatin-like allergen from apple, is highly resistant to gastrointestinal digestion and thermal processing. Int. Arch. Allergy Immunol. 2008, 147, 289–298. [Google Scholar] [CrossRef]
- Fierens, E.; Rombouts, S.; Gebruers, K.; Goesaert, H.; Brijs, K.; Beaugrand, J.; Volckaert, G.; Van Campenhout, S.; Proost, P.; Courtin, C.M.; et al. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family. Biochem. J. 2007, 403, 583–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatters, R.G.; Boykin, L.; Lapointe, S.L.; Hunter, W.B.; Weathersbee, A.A. Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. J. Mol. Evol. 2006, 63, 12–29. [Google Scholar] [CrossRef] [PubMed]
- Abad, L.R.; D’Urzo, M.P.; Liu, D.; Narasimhan, M.L.; Reuveni, M.; Zhu, J.K.; Niu, X.; Singh, N.K.; Hasegawa, P.M.; Bressan, R.A. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996, 118, 11–23. [Google Scholar] [CrossRef]
- Yan, X.; Qiao, H.; Zhang, X.; Guo, C.; Wang, M.; Wang, Y.; Wang, X. Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Cui, J.; Zhou, X.; Luan, Y.; Luan, F. Genome-wide identification, characterization and expression analysis of the TLP gene family in melon (Cucumis melo L.). Genomics 2020, 112, 2499–2509. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Cui, Y.; Qiao, K.; Zhu, L.; Fan, S.; Ma, Q. Comprehensive genome-wide analysis of thaumatin-like gene family in four cotton species and functional identification of GhTLP19 involved in regulating tolerance to verticillium dahlia and drought. Front. Plant Sci. 2020, 11, 575015. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a new osmotin-like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 2017, 8, 410. [Google Scholar] [CrossRef]
- Monteiro, S.; Barakat, M.; Piçarra-Pereira, M.A.; Teixeira, A.R.; Ferreira, R.B. Osmotin and thaumatin from grape: A putative general defense mechanism against pathogenic fungi. Phytopathology 2003, 93, 1505–1512. [Google Scholar] [CrossRef] [Green Version]
- Jayasankar, S.; Li, Z.; Gray, D.J. Constitutive expression of Vitis vinifera thaumatin-like protein after in vitro selection and its role in anthracnose resistance. Funct. Plant Biol. 2003, 30, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Rajam, M.V.; Chandola, N.; Goud, P.S.; Singh, D.; Kashyap, V.; Choudhary, M.L.; Sihachakr, D. Thaumatin gene confers resistance to fungal pathogens as well as tolerance to abiotic stresses in transgenic tobacco plants. Biol. Plant. 2007, 51, 135–141. [Google Scholar] [CrossRef]
- Yasmin, N.; Saleem, M. Biochemical characterization of fruit-specific pathogenesis-related antifungal protein from basrai banana. Microbiol. Res. 2014, 169, 369–377. [Google Scholar] [CrossRef]
- Wang, H.; Ng, T.B. Isolation of an antifungal thaumatin-like protein from kiwi fruits. Phytochemistry 2002, 61, 1–6. [Google Scholar] [CrossRef]
- Mahdavi, F.; Sariah, M.; Maziah, M. Expression of rice thaumatin-like protein gene in transgenic banana plants enhances resistance to fusarium wilt. Appl. Biochem. Biotechnol. 2011, 166, 1008–1019. [Google Scholar] [CrossRef]
- Ojola, P.O.; Nyaboga, E.N.; Njiru, P.N.; Orinda, G. Overexpression of rice thaumatin-like protein (Ostlp) gene in transgenic cassava results in enhanced tolerance to Colletotrichum gloeosporioides f. sp. manihotis. J. Genet. Eng. Biotechnol. 2018, 16, 125–131. [Google Scholar] [CrossRef]
- Singh, N.K.; Kumar, K.R.R.; Kumar, D.; Shukla, P.; Kirti, P.B. Characterization of a pathogen induced thaumatin-like protein gene AdTLP from arachis diogoi, a wild peanut. PLoS ONE 2013, 8, e83963. [Google Scholar] [CrossRef]
- Acharya, K.; Pal, A.K.; Gulati, A.; Kumar, S.; Singh, A.K.; Ahuja, P.S. Overexpression of camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to macrophomina phaseolina and phytophthora infestans infection. Mol. Biotechnol. 2013, 54, 609–622. [Google Scholar] [CrossRef]
- Breiteneder, H. Thaumatin-like proteins—A new family of pollen and fruit allergens. Allergy 2004, 59, 479–481. [Google Scholar] [CrossRef]
- Munis, M.F.H.; Tu, L.; Deng, F.; Tan, J.; Xu, L.; Xu, S.; Long, L.; Zhang, X. A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 2010, 393, 38–44. [Google Scholar] [CrossRef]
- Zhang, T.; Jin, Y.; Zhao, J.-H.; Gao, F.; Zhou, B.; Fang, Y.-Y.; Guo, H.-S. Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol. Plant 2016, 9, 939–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, H.; Chen, W.; Zhao, P.; Gong, H.; Sang, X.; Cui, Y. Regional association analysis-based fine mapping of three clustered QTL for verticillium wilt resistance in cotton (G. hirsutum L.). BMC Genomics 2017, 18, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inderbitzin, P.; Subbarao, K. Verticillium systematics and evolution: How confusion impedes Verticillium wilt management and how to resolve it. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Menu-Bouaouiche, L.; Peumans, W.J.; Payan, F.; Barre, A.; Roussel, A.; Van Damme, E.; Rougé, P. Resolution of the structure of the allergenic and antifungal banana fruit thaumatin-like protein at 1.7-Å. Biochimie 2006, 88, 45–52. [Google Scholar] [CrossRef]
- Theis, T.; Stahl, U. Antifungal proteins: Targets, mechanisms and prospective applications. Cell. Mol. Life Sci. 2004, 61, 437–455. [Google Scholar] [CrossRef]
- Ellegren, H. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 2008, 17, 4586–4596. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Hu, W.; Hou, X.; Huang, C.; Yan, Y.; Tie, W.; Ding, Z.; Wei, Y.; Liu, J.; Miao, H.; Lu, Z.; et al. Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 2015, 16, 19728–19751. [Google Scholar] [CrossRef] [Green Version]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Watanabe, Y. From the cover: Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Willmann, M.R.; Poethig, R.S. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007, 10, 503–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Ho, V.S.; Wong, J.H.; Ng, T. A thaumatin-like antifungal protein from the emperor banana. Peptides 2007, 28, 760–766. [Google Scholar] [CrossRef]
- Garcia-Casado, G.; Collada, C.; Allona, I.; Soto, A.; Casado, R.; Rodriguez-Cerezo, E.; Gomez, L.; Aragoncillo, C. Characterization of an apoplastic basic thaumatin-like protein from recalcitrant chestnut seeds. Physiol. Plant. 2000, 110, 172–180. [Google Scholar] [CrossRef]
- Cao, J.; Lv, Y.; Hou, Z.; Li, X.; Ding, L. Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants. Plant Growth Regul. 2016, 79, 299–307. [Google Scholar] [CrossRef]
- Narasimhan, M.L.; Bressan, R.A.; D’Urzo, M.P.; Jenks, M.A.; Mengiste, T. Unexpected turns and twists in structure/function of PR-proteins that connect energy metabolism and immunity. Adv. Bot. Res. 2009, 51, 439–489. [Google Scholar]
- Wang, Q.; Li, F.; Zhang, X.; Zhang, Y.; Hou, Y.; Zhang, S.; Wu, Z. Purification and characterization of a CkTLP protein from cynanchum komarovii seeds that confers antifungal activity. PLoS ONE 2011, 6, e16930. [Google Scholar] [CrossRef]
- An, M.; Tong, Z.; Ding, C.; Wang, Z.; Sun, H.; Xia, Z.; Qi, M.; Wu, Y.; Liang, Y. Molecular characterization of the thaumatin-like protein PR-NP24 in tomato fruits. J. Agric. Food Chem. 2019, 67, 13001–13009. [Google Scholar] [CrossRef]
- Narasimhan, M.L.; Damsz, B.; Coca, M.; Ibeas, J.; Yun, D.-J.; Pardo, J.M.; Hasegawa, P.M.; Bressan, R.A. A plant defense response effector induces microbial apoptosis. Mol. Cell 2001, 8, 921–930. [Google Scholar] [CrossRef]
- Raghothama, K.G.; Maggio, A.; Narasimhan, M.L.; Kononowicz, A.K.; Wang, G.; D’Urzo, M.P.; Hasegawa, P.M.; Bressan, R.A. Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol. Biol. 1997, 34, 393–402. [Google Scholar] [CrossRef]
- Ramos, M.V.; de Oliveira, R.S.; Pereira, H.; Moreno, F.B.; Lobo, M.D.; Rebelo, L.M.; Brandão-Neto, J.; de Sousa, J.; Monteiro-Moreira, A.C.; Freitas, C.D.; et al. Crystal structure of an antifungal osmotin-like protein from Calotropis procera and its effects on Fusarium solani spores, as revealed by atomic force microscopy: Insights into the mechanism of action. Phytochemistry 2015, 119, 5–18. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, X.; Wang, H.; Yu, T.; Miao, Y.; Zheng, X. Expression of the LePR5 gene from cherry tomato fruit induced by Cryptococcus laurentii and the analysis of LePR5 protein antifungal activity. Postharvest Biol. Technol. 2015, 111, 337–344. [Google Scholar] [CrossRef]
- Freitas, C.D.; Silva, M.Z.; Bruno-Moreno, F.; Monteiro-Moreira, A.C.; Moreira, R.A.; Ramos, M.V. New constitutive latex osmotin-like proteins lacking antifungal activity. Plant Physiol. Biochem. 2015, 96, 45–52. [Google Scholar] [CrossRef]
- Singh, S.; Tripathi, R.K.; Lemaux, P.; Buchanan, B.B.; Singh, J. Redox-dependent interaction between thaumatin-like protein and β-glucan influences malting quality of barley. Proc. Natl. Acad. Sci. USA 2017, 114, 7725–7730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osmond, R.I.W.; Hrmova, M.; Fontaine, F.; Imberty, A.; Fincher, G.B. Binding interactions between barley thaumatin-like proteins and (1,3)-β-D-glucans. JBIC J. Biol. Inorg. Chem. 2001, 268, 4190–4199. [Google Scholar] [CrossRef]
- Zareie, R.; Melanson, D.L.; Murphy, P.J. Isolation of fungal cell wall degrading proteins from barley (Hordeum vulgare L.) leaves infected with Rhynchosporium secalis. Mol. Plant Microbe Interact. 2002, 15, 1031–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Damme, E.J.; Charels, D.; Menu-Bouaouiche, L.; Proost, P.; Barre, A.; Rougé, P.; Peumans, W.J. Biochemical, molecular and structural analysis of multiple thaumatin-like proteins from the elderberry tree (Sambucus nigra L.). Planta 2002, 214, 853–862. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Romanel, E.; Silva, T.F.; Corrêa, R.L.; Farinelli, L.; Hawkins, J.S.; Schrago, C.E.G.; Vaslin, M.F.S. Global alteration of microRNAs and transposon-derived small RNAs in cotton (Gossypium hirsutum) during Cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol. Biol. 2012, 80, 443–460. [Google Scholar] [CrossRef]
- Xie, F.; Sun, G.; Stiller, J.; Zhang, B. Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS ONE 2011, 6, e26980. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Chen, W.; Sang, X.; Wang, T.; Gong, H.; Zhao, Y.; Zhao, P.; Wang, H. Genome-Wide Identification of the Thaumatin-like Protein Family Genes in Gossypium barbadense and Analysis of Their Responses to Verticillium dahliae Infection. Plants 2021, 10, 2647. https://doi.org/10.3390/plants10122647
Zhang Y, Chen W, Sang X, Wang T, Gong H, Zhao Y, Zhao P, Wang H. Genome-Wide Identification of the Thaumatin-like Protein Family Genes in Gossypium barbadense and Analysis of Their Responses to Verticillium dahliae Infection. Plants. 2021; 10(12):2647. https://doi.org/10.3390/plants10122647
Chicago/Turabian StyleZhang, Yilin, Wei Chen, Xiaohui Sang, Ting Wang, Haiyan Gong, Yunlei Zhao, Pei Zhao, and Hongmei Wang. 2021. "Genome-Wide Identification of the Thaumatin-like Protein Family Genes in Gossypium barbadense and Analysis of Their Responses to Verticillium dahliae Infection" Plants 10, no. 12: 2647. https://doi.org/10.3390/plants10122647




