Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses
Abstract
:1. Introduction
2. Results
2.1. Dehydrin Genes of Brachypodium Species and Outgroup Grasses
2.2. Cis-Regulatory Elements of Bdhn Genes
2.3. The Brachypodium Dehydrin Tree
2.4. Chromosome Distributions and Selection Analysis of Duplicated Bdhn Genes
2.5. Dehydrin Gene Clusters, Phylogenetics, and Climate Niche Variation in B. distachyon
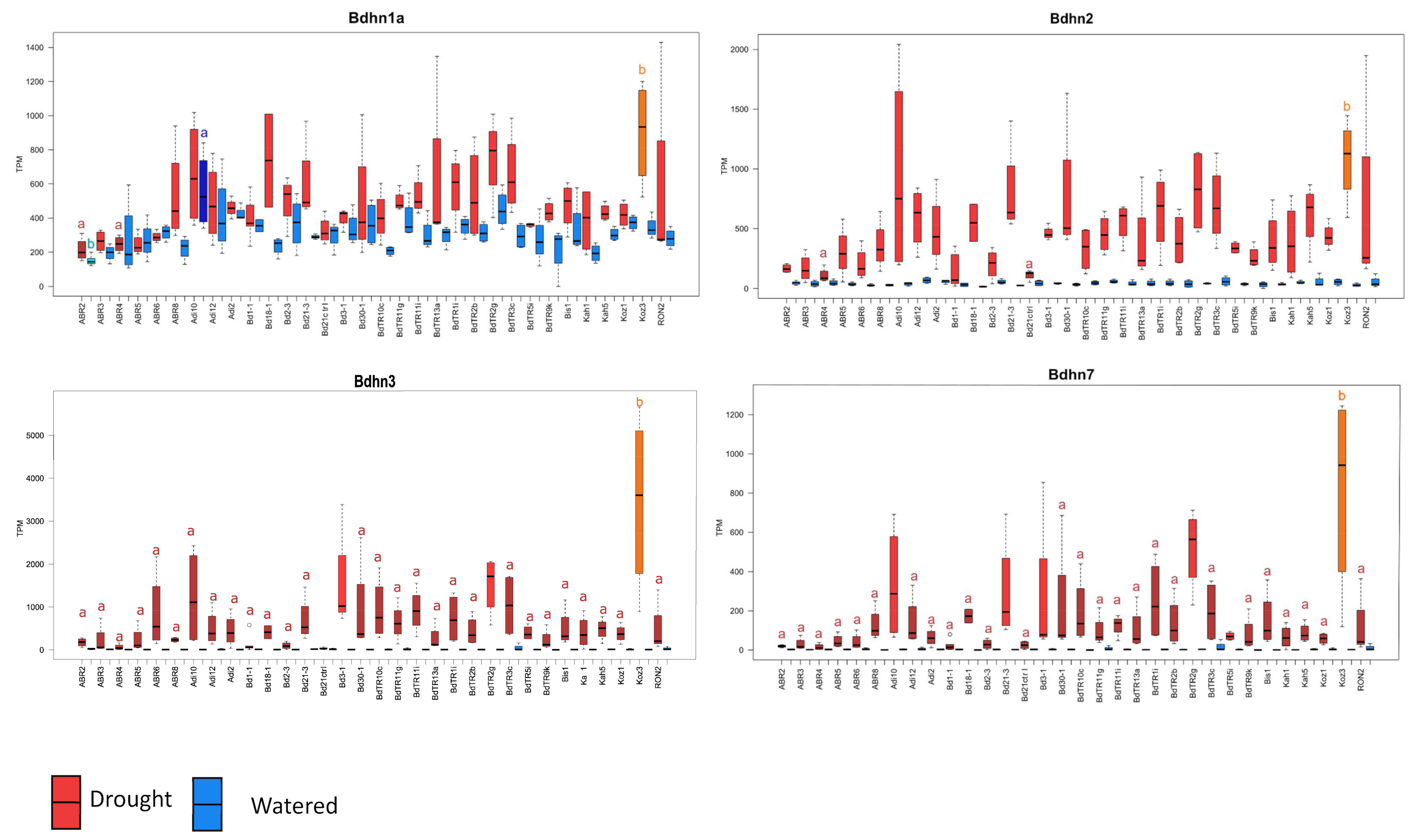
2.6. Differential Expression of Bdhn Genes in Brachypodium distachyon Ecotypes under Drought and Temperature Stress Conditions
2.7. Effects of Drought on Dehydrin Gene Expression and Drought-Response Phenotypic Traits
2.8. Phylogenetic Signal of Dehydrin Expression, Phenotypic Trait Changes and Climate Variation in the Brachypodium distachyon Bdhn Tree
3. Discussion
3.1. The Dehydrin Gene Family in Brachypodium
3.2. Dehydrin Expression Induction in Brachypodium distachyon
3.3. Correlated Dehydrin and Phenotypic Drought Response and Phylogenetic Signal in Brachypodium distachyon
4. Material and Methods
4.1. Identification of Dehydrin Sequences
4.2. Structural Analysis, Conserved Motifs, and Cis-Regulatory Elements (CREs)
4.3. Multiple Alignments and Phylogenetic Analysis
4.4. Chromosomal Location, Gene Duplication, and Selection Analysis
4.5. Clustering and Phylogeny of Dehydrin Genes in Brachypodium distachyon Ecotypes
4.6. Expression Analysis of Dehydrin Genes in Brachypodium distachyon
4.7. Drought-Induced Changes in Dehydrin Expressions, Phenotypic, Physiological, and Climatic Niche Traits, and Phylogenetic Signal in Brachypodium distachyon
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, M.A.; Wani, S.H.; Bhattacharjee, S.; Burritt, D.J.; Tran, L.-S.P. Drought Stress Tolerance in Plants; Springer: Cham, Switzerland, 2016; Volume 2. [Google Scholar]
- Des Marais, D.L.; Lasky, J.R.; Verslues, P.E.; Chang, T.Z.; Juenger, T.E. Interactive effects of water limitation and elevated temperature on the physiology, development and fitness of diverse accessions of Brachypodium distachyon. New Phytol. 2017, 214, 132–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Tommasini, L.; Svensson, J.T.; Rodriguez, E.M.; Wahid, A.; Malatrasi, M.; Kato, K.; Wanamaker, S.; Resnik, J.; Close, T.J. Dehydrin gene expression provides an indicator of low temperature and drought stress: Transcriptome-based analysis of Barley (Hordeum vulgare L.). Funct. Integr. Genom. 2008, 8, 387–405. [Google Scholar] [CrossRef]
- Reddy, S.K.; Liu, S.; Rudd, J.C.; Xue, Q.; Payton, P.; Finlayson, S.A.; Mahan, J.; Akhunova, A.; Holalu, S.V.; Lu, N. Physiology and transcriptomics of water-deficit stress responses in wheat cultivars TAM 111 and TAM 112. J. Plant Physiol. 2014, 171, 1289–1298. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Xu, H.; Zhu, H.; Tao, Y.; Zhang, G.; Zhang, L.; Zhang, C.; Zhang, Z.; Ma, Z. Classification and expression diversification of wheat dehydrin genes. Plant Sci. 2014, 214, 113–120. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and function: A review of the dehydrin protein family. Front. Plant Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, G.; Dhar, Y.V.; Srivastava, D.; Kidwai, M.; Chauhan, P.S.; Bag, S.K.; Asif, M.H.; Chakrabarty, D. Genome-wide analysis of rice dehydrin gene family: Its evolutionary conservedness and expression pattern in response to PEG induced dehydration stress. PLoS ONE 2017, 12, e0176399. [Google Scholar] [CrossRef] [Green Version]
- Riley, A.C.; Ashlock, D.A.; Graether, S.P. Evolution of the modular, disordered stress proteins known as dehydrins. PLoS ONE 2019, 14, e0211813. [Google Scholar] [CrossRef] [Green Version]
- Perdiguero, P.; Collad, C.; Soto, Á. Novel dehydrins lacking complete K-segments in Pinaceae. The exception rather than the rule. Front. Plant Sci. 2014, 5, 682. [Google Scholar] [CrossRef] [Green Version]
- Goday, A.; Jensen, A.B.; Culiáñez-Macià, F.A.; Albà, M.M.; Figueras, M.; Serratosa, J.; Torrent, M.; Pagès, M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 1994, 6, 351–360. [Google Scholar] [CrossRef] [Green Version]
- Rosales, R.; Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. The crucial role of Φ- and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 2014, 108, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Strimbeck, G.R. Hiding in plain sight: The F segment and other conserved features of seed plant SKn dehydrins. Planta 2017, 245, 1061–1066. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, F.; Nicoll, W.S.; Zimmermann, R.; Cheetham, M.E.; Blatch, G.L. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005, 14, 1697–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, A.A.; Veltri, M.; Boddington, K.F.; Singh, K.K.; Graether, S.P. Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Wheat and barley dehydrins under cold, drought, and salinity—What can LEA-II proteins tell us about plant stress response? Front. Plant Sci. 2014, 5, 343. [Google Scholar] [CrossRef] [Green Version]
- Suprunova, T.; Krugman, T.; Fahima, T.; Chen, G.; Shams, I.; Korol, A.; Nevo, E. Differential expression of dehydrin genes in wild barley, Hordeum spontaneum, associated with resistance to water deficit. Plant Cell Environ. 2004, 27, 1297–1308. [Google Scholar] [CrossRef]
- Karami, A.; Shahbazi, M.; Niknam, V.; Shobbar, Z.S.; Tafreshi, R.S.; Abedini, R.; Mabood, H.E. Expression analysis of dehydrin multigene family across tolerant and susceptible barley (Hordeum vulgare L.) genotypes in response to terminal drought stress. Acta Physiol. Plant 2013, 35, 2289–2297. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, X.; Zhang, L. Structural and functional dynamics of dehydrins: A plant protector protein under abiotic stress. Int. J. Mol. Sci. 2018, 19, 3420. [Google Scholar] [CrossRef] [Green Version]
- Lv, A.; Su, L.; Liu, X.; Xing, Q.; Huang, B.; An, Y.; Zhou, P. Characterization of Dehydrin protein, CdDHN4-L and CdDHN4-S, and their differential protective roles against abiotic stress in vitro. BMC Plant Biol. 2018, 18, 299. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Q.; Li, D.; Yang, X.; Li, D. Multifunctional roles of plant dehydrins in response to environmental stresses. Front. Plant Sci. 2017, 8, 1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drira, M.; Saibi, W.; Brini, F.; Gargouri, A.; Masmoudi, K.; Hanin, M. The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 2013, 54, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Liu, Y.; Kong, X.; Liu, Y.; Li, D. Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 2011, 65, 109–118. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.X.; Wei, W.; Han, L.; Guan, Z.Q.; Wang, Z.; Chai, T.Y. BjDHNs confer heavy-metal tolerance in plants. Mol. Biotechnol. 2008, 38, 91–98. [Google Scholar] [CrossRef]
- Hara, M.; Kondo, M.; Kato, T. A KS-type dehydrin and its related domains reduce Cu-promoted radical generation and the histidine residues contribute to the radical-reducing activities. J. Exp. Bot. 2013, 64, 1615–1624. [Google Scholar] [CrossRef] [Green Version]
- Koag, M.C.; Fenton, R.D.; Wilkens, S.; Close, T.J. The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 2003, 131, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, S.L.; Schart, V.; Malcolmson, J.; Hogarth, K.A.; Martynowicz, D.M.; Tralman-Baker, E.; Patel, S.N.; Graether, S.P. The importance of size and disorder in the cryoprotective effects of dehydrins. Plant Physiol. 2013, 163, 1376–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholthof, K.-B.G.; Irigoyen, S.; Catalán, P.; Mandadi, K.K. Brachypodium: A monocot grass model system for plant biology. Plant Cell 2018, 30, 1673–1694. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670. [Google Scholar] [CrossRef]
- Steinwand, M.A.; Young, H.A.; Bragg, J.N.; Tobias, C.M.; Vogel, J.P. Brachypodium sylvaticum, a Model for Perennial Grasses: Transformation and Inbred Line Development. PLoS ONE 2013, 8, e75180. [Google Scholar] [CrossRef]
- Lee, K.; Kang, H. Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 2016, 39, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Ryu, H.; Cho, Y.G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Yoon, J.S.; Kim, J.Y.; Lee, M.B.; Seo, Y.W. Over-expression of the Brachypodium ASR gene, BdASR4, enhances drought tolerance in Brachypodium distachyon. Plant Cell Rep. 2019, 38, 1109–1125. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Ozyigit, I.I.; Tombuloglu, H.; Koc, I. In silico comparative analysis of LEA (Late Embryogenesis Abundant) proteins in Brachypodium distachyon L. Plant Omi. J. 2013, 6, 433–440. [Google Scholar]
- Rodríguez, E.M.; Svensson, J.T.; Malatrasi, M.; Choi, D.W.; Close, T.J. Barley Dhn13 encodes a KS-type dehydrin with constitutive and stress responsive expression. Theor. Appl. Genet. 2005, 110, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 2184. [Google Scholar] [CrossRef] [PubMed]
- Sancho, R.; Cantalapiedra, C.P.; López-Alvarez, D.; Gordon, S.P.; Vogel, J.P.; Catalán, P.; Contreras-Moreira, B. Comparative plastome genomics and phylogenomics of Brachypodium: Flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2018, 218, 1631–1644. [Google Scholar] [CrossRef] [Green Version]
- Sancho, R.; Catalán, P.; Contreras-Moreira, B.; Juenger, T.E.; Des Marais, D.L. Patterns of gene co-expression under water-deficit treatments and pan-genome occupancy in Brachypodium distachyon. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gálvez, S.; Mérida-García, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramírez-González, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Wang, S.; Paetzold, L.; Wang, Z.; Hui, K.; Rudd, J.C.; Xue, Q.; Ibrahim, A.M.H.; Metz, R.; Johnson, C.D.; et al. RNA-seq analysis reveals different drought tolerance mechanisms in two broadly adapted wheat cultivars ‘TAM 111′ and ‘TAM 112’. Sci. Rep. 2021, 11, 4301. [Google Scholar] [CrossRef]
- Catalán, P.; Müller, J.; Hasterok, R.; Jenkins, G.; Mur, L.A.; Langdon, T.; Betekhtin, A.; Siwinska, D.; Pimentel, M.; López-Alvarez, D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 2012, 109, 385–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, D.W.; Zhu, B.; Close, T.J. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 1999, 98, 1234–1247. [Google Scholar] [CrossRef]
- Díaz-Pérez, A.; López-Álvarez, D.; Sancho, R.; Catalán, P. Reconstructing the origins and the biogeography of species ’ genomes in the highly reticulate allopolyploid-rich model grass genus Brachypodium using minimum evolution, coalescence and maximum likelihood approaches. Mol. Phylogenet. Evol. 2018, 127, 256–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Bremont, J.F.; Maruri-López, I.; Ochoa-Alfaro, A.E.; Delgado-Sánchez, P.; Bravo, J.; Rodríguez-Kessler, M. LEA Gene Introns: Is the Intron of Dehydrin Genes a Characteristic of the Serine-Segment? Plant Mol. Biol. Report. 2013, 31, 128–140. [Google Scholar] [CrossRef]
- Martínez, L.M.; Fernández-ocaña, A.; Rey, P.J.; Salido, T.; Amil-ruiz, F.; Manzaneda, A.J. Variation in functional responses to water stress and differentiation between natural allopolyploid populations in the Brachypodium distachyon species complex. Ann. Bot. 2018, 121, 1369–1382. [Google Scholar] [CrossRef]
- Martínez, L.M. Análisis de la Variación Genética, Respuesta Funcional y Expresión Génica Frente a Diferentes Tipos de Estrés en el Complejo de Ploidía Brachypodium Distachyon (Poaceae); Universidad de Jaén: Jaén, Spain, 2020. [Google Scholar]
- Fisher, L.H.; Han, J.; Corke, F.M.; Akinyemi, A.; Didion, T.; Nielsen, K.K.; Doonan, J.H.; Mur, L.A.; Bosch, M. Linking Dynamic Phenotyping with Metabolite Analysis to Study Natural Variation in Drought Responses of Brachypodium distachyon. Front. Plant Sci. 2016, 7, 1751. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [Green Version]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [Green Version]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef] [Green Version]
- Cernusak, L.A.; Ubierna, N.; Winter, K.; Holtum, J.A.M.; Marshall, J.D.; Farquhar, G.D. Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 2013, 200, 950–965. [Google Scholar] [CrossRef] [PubMed]
- Manzaneda, A.J.; Rey, P.J.; Anderson, J.T.; Raskin, E.; Weiss-Lehman, C.; Mitchell-Olds, T. Natural variation, differentiation, and genetic trade-offs of ecophysiological traits in response to water limitation in Brachypodium distachyon and its descendent allotetraploid B. hybridum (Poaceae). Evolution 2015, 69, 2689–2704. [Google Scholar] [CrossRef] [Green Version]
- Revell, L.J.; Harmon, L.J.; Collar, D.C. Phylogenetic signal, evolutionary process, and rate. Syst. Biol. 2008, 57, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ding, Y.; Zhu, C. Sensitivity and Responses of Chloroplasts to Heat Stress in Plants. Front. Plant Sci. 2020, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Contreras-Moreira, B.; De Silva, N.; Maslen, G.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Barba, M.; Bolser, D.M.; Cambell, L.; et al. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020, 48, D689–D695. [Google Scholar] [CrossRef] [Green Version]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; McPartlon, M.; Li, J. Improved protein structure prediction by deep learning irrespective of co-evolution information. Nat. Mach. Intell. 2021, 3, 601–609. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. ICn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics 2020, 36, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.T.T.; Contreras-Moreira, B.; Castro-Mondragon, J.A.; Santana-Garcia, W.; Ossio, R.; Robles-Espinoza, C.D.; Bahin, M.; Collombet, S.; Vincens, P.; Thieffry, D.; et al. RSAT 2018: Regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res. 2018, 46, W209–W214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ksouri, N.; Castro-Mondragón, J.A.; Montardit-Tarda, F.; van Helden, J.; Contreras-Moreira, B.; Gogorcena, Y. Tuning promoter boundaries improves regulatory motif discovery in nonmodel plants: The peach example. Plant Physiol. 2021, 185, 1242–1258. [Google Scholar] [CrossRef]
- Castro-Mondragon, J.A.; Jaeger, S.; Thieffry, D.; Thomas-Chollier, M.; Van Helden, J. RSAT matrix-clustering: Dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res. 2017, 45, e119. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, S.; Jørgensen, B.; Aziz, E.; Batool, R.; Naseer, S.; Rasmussen, S.K. Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley. Plants 2020, 9, 1439. [Google Scholar] [CrossRef]
- Smith, M.D.; Wertheim, J.O.; Weaver, S.; Murrell, B.; Scheffler, K.; Kosakovsky Pond, S.L. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015, 32, 1342–1353. [Google Scholar] [CrossRef] [Green Version]
- Murrell, B.; Weaver, S.; Smith, M.D.; Wertheim, J.O.; Murrell, S.; Aylward, A.; Eren, K.; Pollner, T.; Martin, D.P.; Smith, D.M.; et al. Gene-wide identification of episodic selection. Mol. Biol. Evol. 2015, 32, 1365–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosakovsky Pond, S.L.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef]
- IBI Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [CrossRef]
- Konstantinov, D.K.; Zubairova, U.S.; Ermakov, A.A.; Doroshkov, A.V. Comparative transcriptome profiling of a resistant vs. susceptible bread wheat (Triticum aestivum L.) cultivar in response to water deficit and cold stress. PeerJ 2021, 9, e11428. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 2018, 19, 111. [Google Scholar] [CrossRef] [PubMed]
- Dray, S.; Dufour, A.-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4b10; Sinauer Associates: Sunderland, MA, USA, 2003; ISBN 0022-2844. [Google Scholar]
- Blomberg, S.P.; Garland, T.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M. Inferring the historical patterns of biological evolution. Nature 1999, 401, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Revell, L.J. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]






| Bdhn | Structure | Panther Family | Panther Subfamily | Phytozome Dehydrin Gene Codes | ||||
|---|---|---|---|---|---|---|---|---|
| B. distachyon (Bd21) | B. stacei (ABR114) | B. hybridum D (ABR113) | B. hybridum S (ABR113) | B. sylvaticum (Ain1) | ||||
| Bdhn1a | PTH33346 | ERD14 | Bradi5g10860 | Brast09G089800 | Brahy.D05G0138300 | Brahy.S09G0091400 | Brasy9G143400 | |
| Bdhn1b | FSKn | Brasy9G149400 | ||||||
| Bdhn2 | FSKn | Bradi3g51200 | Brast04G110500 | Brahy.D03G0707100 | Brahy.S04G0117200 | Brasy4G117200 | ||
| Bdhn3 | YnSKn | SF23 | Bradi1g37410 | Brast07G152200 | Brahy.D01G0507100 | Brahy.S07G0169500 | Brasy7G144000 | |
| Bdhn4 | YnSKn | SF14 | Bradi4g22280 | |||||
| Bdhn5 | YnSKn | Bradi4g22290 | Brast05G049400 | Brahy.D04G0319400 | Brahy.S05G0054400 | Brasy5G271500 | ||
| Bdhn6 | YnSKn | SF 19 | Bradi4g19525 | Brast05G075400 | Brahy.D04G0277700 | Brahy.S05G0083200 | Brasy4G237500 | |
| Bdhn7 | YnSKn | SF14 | Bradi3g43870 | Brast04G194300 | Brahy.D03G0604200 | Brahy.S04G0208500 | Brasy4G220100 | |
| Bdhn8 | YnSKn | Bradi3g43855 | Brast04G197200 | Brahy.D03G0604000 | Brahy.S04G0208900 | Brasy4G219800 | ||
| Bdhn9 | YnSKn | XERO I | Bradi2g47575 | Brast01G171900 | Brahy.D02G0637300 | Brahy.S01G0182800 | Brasy1G228900 | |
| Bdhn10 | K*NLSL-S | PTHR34941 | HIRD 11 | Bradi1g13330 | Brast02G251900 | Brahy.D01G017200 | Brahy.S02G0268100 | Brasy2G277500 |
| Family ID | Motif ID | Ncor | Sig | TF Binding Site | Bdhn Genes (No. Sites Found) |
|---|---|---|---|---|---|
| BRI1-EMS suppressor/brassinazole-resistant | BES1/BZR1 | 0.719 | 4.08 |  | BD: Bdhn5(1) Bdhn6(1) Bdhn7(3) Bdhn9(1) |
| BHD: Bdhn1(1) Bdhn2(2) Bdhn5(1) Bdhn6(1) Bdhn7(3) Bdhn9(1) | |||||
| BHS: Bdhn1(1) Bdhn2(2) Bdhn5(4) Bdhn6(1) Bdhn7(2) Bdhn8(1) | |||||
| BS: Bdhn1(1) Bdhn2(2) Bdhn5(3) Bdhn6(1) Bdhn7(2) Bdhn8(1) | |||||
| BSY: Bdhn2(1) Bdhn5(1) Bdhn7(1) Bdhn8(2) Bdhn9(1) | |||||
| Myb | MYB124 | 0.598 | 2.18 |  | BD: Bdhn1(5) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) Bdhn8(2) |
| BHD: Bdhn1(7) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) Bdhn8(2) | |||||
| BHS: Bdhn1(7) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn8(2) | |||||
| BS: Bdhn1(8) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn8(2) | |||||
| BSY: Bdhn1a(4) Bdhn1b(4) Bdhn2(4) Bdhn3(2) Bdhn6(1) Bdhn7(2) | |||||
| C2H2 zinc finger | ZAT | 0.738 | 4.91 |  | BD: Bdhn4(3) Bdhn5(1) Bdhn10(2) |
| BHD: Bdhn5(1) | |||||
| BHS: Bdhn5(1) Bdhn10(1) | |||||
| BS: Bdhn5(2) Bdhn10(1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decena, M.A.; Gálvez-Rojas, S.; Agostini, F.; Sancho, R.; Contreras-Moreira, B.; Des Marais, D.L.; Hernandez, P.; Catalán, P. Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants 2021, 10, 2664. https://doi.org/10.3390/plants10122664
Decena MA, Gálvez-Rojas S, Agostini F, Sancho R, Contreras-Moreira B, Des Marais DL, Hernandez P, Catalán P. Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants. 2021; 10(12):2664. https://doi.org/10.3390/plants10122664
Chicago/Turabian StyleDecena, Maria Angeles, Sergio Gálvez-Rojas, Federico Agostini, Ruben Sancho, Bruno Contreras-Moreira, David L. Des Marais, Pilar Hernandez, and Pilar Catalán. 2021. "Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses" Plants 10, no. 12: 2664. https://doi.org/10.3390/plants10122664
APA StyleDecena, M. A., Gálvez-Rojas, S., Agostini, F., Sancho, R., Contreras-Moreira, B., Des Marais, D. L., Hernandez, P., & Catalán, P. (2021). Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants, 10(12), 2664. https://doi.org/10.3390/plants10122664









