An Improved Method for the Extraction of Nucleic Acids from Plant Tissue without Grinding to Detect Plant Viruses and Viroids
Abstract
:1. Introduction
2. Results
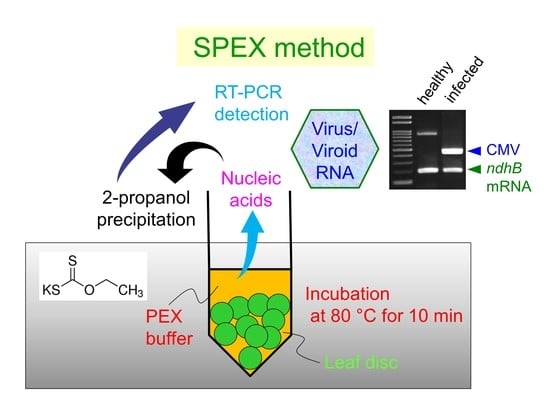
2.1. Incubation Conditions of Leaf Discs with PEX Buffer
2.2. Quality and Quantity of Extracted Nucleic Acids
2.3. Nucleic Acid Extraction from Preserved Leaves Using the SPEX Method
2.4. Application of the SPEX Method for the Detection of Pathogens from Crops
3. Discussion
4. Materials and Methods
4.1. Plant Virus and Viroid Sources and Plant Growth Conditions
4.2. Simplified Extraction of Nucleic Acids Using PEX Buffer
4.3. Differential Precipitation of Nucleic Acids with 2-Butoxyethanol
4.4. RT-PCR
4.5. Quantity and Quality Evaluation of Extracted Nucleic Acids
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yvon, M.; Vile, D.; Brault, V.; Blanc, S.; van Munster, M. Drought reduces transmission of turnip yellows virus, an insect-vectored circulative virus. Virus Res. 2017, 241, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Navarro, B.; Flores, R.; Di Serio, F. Advances in viroid-host interactions. Annu. Rev. Virol. 2021, 8, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Pallás, V.; Sánchez-Navarro, J.A.; James, D. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 2018, 9, 2087. [Google Scholar] [CrossRef] [Green Version]
- Kwok, S.; Higuchi, R. Avoiding false positives with PCR. Nature 1989, 339, 237–238. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.H. Optimization and troubleshooting in PCR. Genome Res. 1995, 4, S185–S194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Champlot, S.; Berthelot, C.; Pruvost, M.; Bennett, E.A.; Grange, T.; Geigl, E.-M. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE 2010, 5, e13042. [Google Scholar] [CrossRef] [Green Version]
- Vandewoestyne, M.; van Hoofstat, D.; de Groote, S.; van Thuyne, N.; Haerinck, S.; van Nieuwerburgh, F.; Deforce, D. Sources of DNA contamination and decontamination procedures in the forensic laboratory. J. Forensic Res. 2011, S2. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Renevey, N.; Thür, B.; Hoffmann, D.; Beer, M.; Hoffmann, B. Efficacy assessment of nucleic acid decontamination reagents used in molecular diagnostic laboratories. PLoS ONE 2016, 11, e0159274. [Google Scholar] [CrossRef]
- Jhingan, A.K. A novel technology for DNA isolation. Methods Mol. Cell Biol. 1992, 3, 15–22. [Google Scholar]
- Williams, C.E.; Ronald, P.C. PCR template-DNA isolated quickly from monocot and dicot leaves without tissue homogenization. Nucleic Acids Res. 1994, 22, 1917–1918. [Google Scholar] [CrossRef] [Green Version]
- Nakahara, K.; Hataya, T.; Uyeda, I. A simple, rapid method of nucleic acid extraction without tissue homogenization for detecting viroids by hybridization and RT-PCR. J. Virol. Methods 1999, 77, 47–58. [Google Scholar] [CrossRef]
- Thompson, J.R.; Wetzel, S.; Klerks, M.M.; Vašková, D.; Schoen, C.D.; Špak, J.; Jelkmann, W. Multiplex RT-PCR detection of four aphid-borne strawberry viruses in Fragaria spp. in combination with a plant mRNA specific internal control. J. Virol. Methods 2003, 111, 85–93. [Google Scholar] [CrossRef]
- Hataya, T. Duplex reverse transcription-polymerase chain reaction system to detect potato spindle tuber viroid using an internal control mRNA and a non-infectious positive control RNA. J. Gen. Plant Pathol. 2009, 75, 167–172. [Google Scholar] [CrossRef]
- Koetsier, G.; Cantor, E. A Practical Guide to Analyzing Nucleic Acid Concentration and Purity with Microvolumes Spectrophotometers. New England Biolabs–Technical Note. 2019. Available online: https://www.bioke.com/blobs/downloads/NEB/MVS_Analysis_of_NA_Concentration_and_Purity.pdf (accessed on 4 December 2021).
- Hataya, T.; Uchino, K.; Arimoto, R.; Suda, N.; Sano, T.; Shikata, E.; Uyeda, I. Molecular characterization of hop latent virus and phylogenetic relationships among viruses closely related to carlaviruses. Arch. Virol. 2000, 145, 2503–2524. [Google Scholar] [CrossRef] [PubMed]
- Hataya, T.; Hikage, K.; Suda, N.; Nagata, T.; Li, S.; Itoga, Y.; Tanikoshi, T.; Shikata, E. Detection of hop latent viroid (HLVd) using reverse transcription and polymerase chain reaction (RT-PCR). Ann. Phytopath. Soc. Jpn. 1992, 58, 677–684. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Kanematsu, S.; Koganezawa, H.; Tsuchizaki, T.; Yoshida, K. Detection of a viroid associated with apple fruit crinkle disease. Ann. Phytopath. Soc. Jpn. 1993, 59, 520–527. [Google Scholar] [CrossRef]
- Sano, T.; Yoshida, H.; Goshono, M.; Monma, T.; Kawasaki, H.; Ishizaki, K. Characterization of a new viroid strain from hops: Evidence for viroid speciation by isolation in different host species. J. Gen. Plant Pathol. 2004, 70, 181–187. [Google Scholar] [CrossRef]
- Nakaune, R.; Nakano, M. Identification of a new apscaviroid from Japanese persimmon. Arch. Virol. 2008, 153, 969–972. [Google Scholar] [CrossRef]
- Nakahara, K.; Hataya, T.; Uyeda, I.; Ieki, H. An improved procedure for extracting nucleic acids from citrus tissues for diagnosis of citrus viroids. Ann. Phytopathol. Soc. Jpn. 1998, 64, 532–538. [Google Scholar] [CrossRef]
- Sipahioglu, H.M.; Usta, M.; Ocak, M. Use of dried high-phenolic laden host leaves for virus and viroid preservation and detection by PCR methods. J. Virol. Methods 2006, 137, 120–124. [Google Scholar] [CrossRef]
- Naoi, T.; Hataya, T. Tolerance even to lethal strain of potato spindle tuber viroid found in wild tomato species can be introduced by crossing. Plants 2021, 10, 575. [Google Scholar] [CrossRef] [PubMed]
- Hataya, T.; Naoi, T. Precisely monomeric linear RNAs of viroids belonging to Pospiviroid and Hostuviroid genera are infectious regardless of transcription initiation site and 5’-terminal structure. Cells 2021, 10, 2971. [Google Scholar] [CrossRef] [PubMed]
- De Ungria, M.C.A.; Tillett, D.; Neilan, B.A.; Cox, P.T.; Lee, A. A novel method of extracting plasmid DNA from Helicobacter species. Helicobacter 1998, 3, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Tillet, D.; Neilan, B.A. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 2000, 36, 251–258. [Google Scholar] [CrossRef]
- Leuko, S.; Goh, F.; Ibáñez-Peral, R.; Burns, B.P.; Walter, M.R.; Neilan, B.A. Lysis efficiency of standard DNA extraction methods for Halococcus spp. in an organic rich environment. Extremophiles 2008, 12, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Phlips, E.J. Improved methods for the isolation of cyanobacterial DNA from environmental samples. J. Phycol. 2009, 45, 517–521. [Google Scholar] [CrossRef]
- Singh, O.A.; Gunapati, O.; Singh, O.K.; Tiwari, O.N. Isolation of fresh water cyanobacterial DNA of north east India by modified xanthogenate method. Int. J. Res. BioSci. 2013, 2, 75–82. [Google Scholar]
- Judelson, H.S.; Messenger-Routh, B. Quantitation of Phytophthora cinnamomi in avocado roots using a species-specific DNA probe. Phytopathology 1996, 86, 763–768. [Google Scholar] [CrossRef]
- Martin, K.J.; Rygiewicz, P.T. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Hataya, T.; Nakahara, K.; Furuta, K.; Shikata, E. Comparison of gene diagnostic methods for the practical diagnosis of chrysanthemum stunt viroid in chrysanthemum plants. Arch. Phytopath. Pflanz. 1999, 32, 179–192. [Google Scholar] [CrossRef]
- Ikegami, H.; Koshita, Y.; Yakushiji, H.; Hirashima, K.; Hirata, C.; Nakahara, T. Simple and efficient RNA extraction and gene analysis in vegetative organs of Japanese persimmon. Plant Biotechnol. 2009, 26, 427–429. [Google Scholar] [CrossRef] [Green Version]
- Hataya, T.; Inoue, A.K.; Ohshima, K.; Shikata, E. Characterization and strain identification of a potato virus Y isolate non-reactive with monoclonal antibodies specific to the ordinary and necrotic strains. Intervirology 1994, 37, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hataya, T.; Furuta, K.; Horita, H.; Sano, T.; Shikata, E. Occurrence of chrysanthemum stunt disease in Hokkaido and detection of chrysanthemum stunt viroid by electrophoresis and hybridization. Ann. Rept. Plant Prot. North Jpn. 1997, 48, 113–117. [Google Scholar]
- Sano, T.; Hataya, T.; Sasaki, A.; Shikata, E. Etrog citron is latently infected with hop stunt viroid-like RNA. Proc. Jpn. Acad. 1986, 62B, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Hataya, T.; Inoue, A.K.; Shikata, E. A PCR-microplate hybridization method for plant virus detection. J. Virol. Methods 1994, 46, 223–236. [Google Scholar] [CrossRef]






| Exp. | Target | Standard PEX | SPEX-A | SPEX-B | SPEX-C |
|---|---|---|---|---|---|
| 1st (10 dpi) | CMV | +++ | (+) | +++ | +++ |
| ndhB mRNA | + | − | ++ | + | |
| 2nd (14 dpi) | CMV | + | +++ | +++ | +++ |
| ndhB mRNA | − | ++ | ++ | ++ | |
| 3rd (10 dpi) | CMV | +++ | +++ | +++ | +++ |
| ndhB mRNA | + | + | + | + | |
| 4th (14 dpi) | CMV | + | + | +++ | +++ |
| ndhB mRNA | + | − | ++ | ++ | |
| 5th (14 dpi) | CMV | +++ | ++ | +++ | +++ |
| ndhB mRNA | ++ | + | ++ | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hataya, T. An Improved Method for the Extraction of Nucleic Acids from Plant Tissue without Grinding to Detect Plant Viruses and Viroids. Plants 2021, 10, 2683. https://doi.org/10.3390/plants10122683
Hataya T. An Improved Method for the Extraction of Nucleic Acids from Plant Tissue without Grinding to Detect Plant Viruses and Viroids. Plants. 2021; 10(12):2683. https://doi.org/10.3390/plants10122683
Chicago/Turabian StyleHataya, Tatsuji. 2021. "An Improved Method for the Extraction of Nucleic Acids from Plant Tissue without Grinding to Detect Plant Viruses and Viroids" Plants 10, no. 12: 2683. https://doi.org/10.3390/plants10122683