Evolutionary Analysis of the YABBY Gene Family in Brassicaceae
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of YABBY Genes in 37 Brassicaceae Species
2.2. Phylogenetic Analysis of YABBY Genes among Brassicaceae Species
2.3. Phylogenetic Relationships among YABBY Genes of U’s Triangle Brassica Diploid and Allotetraploid Species
2.4. Syntenic Relationships among YABBY Genes of Different Brassicaceae Species
2.5. Putative cis-Regulatory Element Analysis of B. rapa and B. oleracea YABBY Genes
2.6. Expression Analysis of YABBY Genes in B. rapa and B. oleracea
3. Discussion
4. Materials and Methods
4.1. Identification of YABBY Protein Genes
4.2. Phylogenetic Analysis
4.3. Syntenic Relationships
4.4. Promoter Region Analysis
4.5. Expression Analysis of YABBY Genes in B. rapa and B. oleracea
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bartholmes, C.; Hidalgo, O.; Gleissberg, S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. 2012, 14, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Floyd, S.K.; Conway, S.J.; Zhong, B.; Scutt, C.P.; Bowman, J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016, 18, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Romanova, M.A.; Maksimova, A.I.; Pawlowski, K.; Voitsekhovskaja, O.V. YABBY Genes in the Development and Evolution of Land Plants. Int. J. Mol. Sci. 2021, 22, 4139. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc-finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar] [CrossRef]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc-finger and HMG-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef]
- Sawa, S.; Ito, T.; Shimura, Y.; Okada, K. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 1999, 11, 69–86. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, J.M.; Broadhvest, J.; Hauser, B.A.; Meister, R.J.; Schneitz, K.; Gasser, C.S. INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules. Genes Dev. 1999, 13, 3160–3169. [Google Scholar] [CrossRef] [Green Version]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef] [Green Version]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y. Bowman, JL. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Atkinson, A.; Otsuga, D.; Christensen, T.; Reynolds, L.; Drews, G.N. The Arabidopsis FILAMENTOUS FLOWER gene is required for FLOWER formation. Development 1999, 126, 2715–2726. [Google Scholar] [CrossRef]
- Kumaran, M.K.; Ye, D.; Yang, W.C.; Griffith, M.E.; Chaudhury, A.M.; Sundaresan, V. Molecular cloning of ABNORMAL FLORAL ORGANS: A gene required for flower development in Arabidopsis. Sex Plant Reprod. 1999, 12, 118–122. [Google Scholar] [CrossRef]
- Lugassi, N.; Nakayama, N.; Bochnik, R.; Zik, M. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 2010, 10, 1–13. [Google Scholar] [CrossRef]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Tanaka, W.; Toriba, T.; Ohmori, Y.; Yoshida, A.; Kawai, A.; Mayama-Tsuchida, T.; Ichikawa, H.; Mitsuda, N.; Ohme-Takagi, M.; Hirano, H.Y. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell 2012, 24, 80–95. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, W.; Toriba, T.; Hirano, H.Y. Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol. 2017, 215, 825–839. [Google Scholar] [CrossRef] [Green Version]
- Dai, M.; Zhao, Y.; Ma, Q.; Hu, Y.; Hedden, P.; Zhang, Q.; Zhou, D.X. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007, 144, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.; Hur, J.; Kim, S.J.; Han, M.J.; Kim, S.R.; An, G. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol. Biol. 2004, 56, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Hu, Y.; Zhao, Y.; Liu, H.; Zhou, D.X. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007, 144, 380–390. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.L.; Xu, Y.Y.; Xu, Z.H.; Chong, K. A rice YABBY gene, OsYABBY4, preferentially expresses in developing vascular tissue. Dev. Genes Evol. 2007, 217, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, Y.; Li, J. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J. Exp. Bot. 2016, 67, 5545–5556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagasawa, N.; Miyoshi, M.; Sano, Y.; Satoh, H.; Hirano, H.Y.; Sakai, H.; Nagato, Y. SUPERWOMAN 1 and DROOPING LEAF genes control floral organ identity in rice. Development 2003, 130, 705–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.Y. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmori, Y.; Toriba, T.; Nakamura, H.; Ichikawa, H.; Hirano, H.Y. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. Plant J. 2011, 65, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Su, H.Y.; Song, J.; Zhao, X.Y.; Zhao, W.; Zhang, X.S. Ectopic expression of TaYAB1, a member of YABBY gene family in wheat, causes the partial abaxialization of the adaxial epidermises of leaves and arrests the development of shoot apical meristem in Arabidopsis. Plant Sci. 2006, 170, 364–371. [Google Scholar] [CrossRef]
- Yang, C.J.; Kursel, L.E.; Studer, A.J.; Bartlett, M.E.; Whipple, C.J.; Doebley, J.F. A gene for genetic background in Zea mays: Fine-mapping enhancer of teosinte branched1.2 to a YABBY class transcription factor. Genetics 2016, 204, 1573–1585. [Google Scholar] [CrossRef] [Green Version]
- Strable, J.; Wallace, J.G.; Unger-Wallace, E.; Briggs, S.; Bradbury, P.J.; Buckler, E.S.; Vollbrecht, E. Maize YABBY Genes drooping leaf1 and drooping leaf2 regulate plant architecture. Plant Cell 2017, 29, 1622–1641. [Google Scholar] [CrossRef] [Green Version]
- Strable, J.; Vollbrecht, E. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development 2019, 146, 171181. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Li, X.; Shannon, L.M.; Yeh, C.T.; Wang, M.L.; Bai, G.; Peng, Z.; Li, J.; Trick, H.N.; Clemente, T.E.; et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012, 44, 720–724. [Google Scholar] [CrossRef] [Green Version]
- Golz, J.F.; Roccaro, M.; Kuzoff, R.; Hudson, A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 2004, 131, 3661–3670. [Google Scholar] [CrossRef] [Green Version]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800. [Google Scholar] [CrossRef]
- Orashakova, S.; Lange, M.; Lange, S.; Wege, S.; Becker, A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 2009, 58, 682–693. [Google Scholar] [CrossRef]
- Lora, J.; Hormaza, J.I.; Herrero, M.; Gasser, C.S. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 2011, 108, 5461–5465. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, L.; Sun, X.; Li, Y.; Yao, J.; van Nocker, S.; Wang, X. Genome-wide analysis of the YABBY gene family in grapevine and functional characterization of VvYABBY4. Front. Plant Sci. 2019, 10, 1207. [Google Scholar] [CrossRef]
- di Rienzo, V.; Imanifard, Z.; Mascio, I.; Gasser, C.S.; Skinner, D.J.; Pierri, C.L.; Marini, M.; Fanelli, V.; Sabetta, W.; Montemurro, C.; et al. Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation. Hortic. Res. 2021, 8, 29. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yang, Z.P.; Zhang, J.; Zhang, L.G. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 2013, 14, 14872–14891. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.; Lin, Y.; Hou, X. Ectopic expression of a pak-choi YABBY Gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana. Genes 2020, 11, 370. [Google Scholar] [CrossRef] [Green Version]
- Toriba, T.; Harada, K.; Takamura, A.; Nakamura, H.; Ichikawa, H.; Suzaki, T.; Hirano, H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007, 277, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Van Houten, J.; Gonzalez, G.; Xiao, H.; van der Knaap, E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013, 288, 111–129. [Google Scholar] [CrossRef] [PubMed]
- İnal, B.; Büyük, İ.; İlhan, E.; Aras, S. Genome-wide analysis of Phaseolus vulgaris C2C2-YABBY transcription factors under salt stress conditions. 3 Biotech 2017, 5, 302. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.P.; Lu, D.; Yu, T.F.; Ji, Y.J.; Zheng, W.J.; Zhang, S.X.; Chai, S.C.; Chen, Z.Y.; Cui, X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017, 119, 132–146. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Wang, L.; Jin, Y.; Xi, J.; Li, Z.; Qin, W.; Yang, Z.; Lu, L.; Chen, Q. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 2018, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, G.; Cai, M.; Priyadarshani, S.; Aslam, M.; Zhou, Q.; Huang, X.; Wang, X.; Liu, Y.; Qin, Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int. J. Mol. Sci. 2019, 20, 5863. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Liu, C.; Ge, D.; Yan, M.; Ren, Y.; Huang, X.; Yuan, Z. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 2020, 752, 144784. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Hsiao, Y.Y.; Chang, S.B.; Zhang, D.; Lan, S.R.; Liu, Z.J.; Tsai, W.C. Genome-wide identification of YABBY genes in orchidaceae and their expression patterns in Phalaenopsis orchid. Genes 2020, 11, 955. [Google Scholar] [CrossRef]
- Liu, X.; Liao, X.Y.; Zheng, Y.; Zhu, M.J.; Yu, X.; Jiang, Y.T.; Zhang, D.; Ma, L.; Xu, X.Y.; Liu, Z.J.; et al. Genome-wide identification of the YABBY gene family in seven species of magnoliids and expression analysis in Litsea. Plants 2020, 10, 21. [Google Scholar] [CrossRef]
- Yamada, T.; Yokota, S.; Hirayama, Y.; Imaichi, R.; Kato, M.; Gasser, C.S. Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 2011, 67, 26–36. [Google Scholar] [CrossRef]
- Talalay, P.; Fahey, J.W. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 2001, 131, 3027–3033. [Google Scholar] [CrossRef]
- Kayaçetin, F.; Efeoğlu, B.; Sarıoğlu, G. Evaluation of fatty acid compositions of some important wild and domestic Turkish mustard genotypes (Brassica spp.). Int. J. Second. Metab. 2018, 5, 270–278. [Google Scholar] [CrossRef]
- Koch, M.A.; German, D.A.; Kiefer, M.; Franzke, A. Database taxonomics as key to modern plant biology. Trends Plant Sci. 2018, 23, 4–6. [Google Scholar] [CrossRef]
- Beilstein, M.A.; Al-Shehbaz, I.A.; Kellogg, E.A. Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 2006, 93, 607–619. [Google Scholar] [CrossRef]
- Beilstein, M.A.; Al-Shehbaz, I.A.; Mathews, S.; Kellogg, E.A. Brassicaceae phylogeny inferred from phytochrome A and ndhF data: Tribes and trichomes revisited. Am. J. Bot. 2008, 95, 1307–1327. [Google Scholar] [CrossRef] [Green Version]
- Franzke, A.; Lysak, M.A.; Al-Shehbaz, I.A.; Koch, M.A.; Mummenhoff, K. Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 2011, 16, 108–116. [Google Scholar] [CrossRef]
- Huang, C.H.; Sun, R.; Hu, Y.; Zeng, L.; Zhang, N.; Cai, L.; Zhang, Q.; Koch, M.A.; Al-Shehbaz, I.; Edger, P.P.; et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 2016, 33, 394–412. [Google Scholar] [CrossRef] [Green Version]
- Nikolov, L.A.; Shushkov, P.; Nevado, B.; Gan, X.; Al-Shehbaz, I.A.; Filatov, D.; Bailey, C.D.; Tsiantis, M. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 2019, 222, 1638–1651. [Google Scholar] [CrossRef] [Green Version]
- Morinaga, T. Interspecific hybridization in Brassica I. The cytology of F1 hybrids of B. napella and various other species with 10 chromosomes. Cytologia 1929, 1, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Schranz, M.E. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 2006, 18, 1152–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lysak, M.A.; Cheung, K.; Kitschke, M.; Bureš, P. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 2007, 145, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Koch, M.A.; Pecinka, A.; Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genom. Res. 2005, 15, 516–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, A.K.; Garza, E.R.; Dietz, V.A.; Hernandez, O.J.; Hawkins, W.D.; Burrell, A.M.; Pepper, A.E. Transcriptome signatures of selection, drift, introgression, and gene duplication in the evolution of an extremophile endemic plant. Genom. Biol. Evol. 2017, 9, 3478–3494. [Google Scholar] [CrossRef] [Green Version]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef] [Green Version]
- Kagale, S.; Robinson, S.J.; Nixon, J.; Xiao, R.; Huebert, T.; Condie, J.; Kessler, D.; Clarke, W.E.; Edger, P.P.; Links, M.G.; et al. Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 2014, 26, 2777–2791. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Feist, L.J.; Parker, D.R. Ecotypic variation in selenium accumulation among populations of Stanleya pinnata. New Phytol. 2001, 149, 61–69. [Google Scholar] [CrossRef]
- Freeman, J.L.; Banuelos, G.S. Selection of salt and boron tolerant selenium hyperaccumulator Stanleya pinnata genotypes and characterization of Se phytoremediation from agricultural drainage sediments. Environ. Sci. Technol. 2011, 45, 9703–9710. [Google Scholar] [CrossRef]
- Cacho, I.N.; Burrell, A.M.; Pepper, A.E.; Strauss, S.Y. Novel nuclearmarkers inform the systematics and the evolution of serpentine use in Streptanthus and allies (Thelypodieae, Brassicaceae). Mol. Phylogenet. Evol. 2014, 72, 71–81. [Google Scholar] [CrossRef]
- Walden, N.; German, D.A.; Wolf, E.M.; Kiefer, M.; Rigault, P.; Huang, X.C.; Kiefer, C.; Schmickl, R.; Franzke, A.; Neuffer, B.; et al. Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae. Nat. Commun. 2020, 11, 3795. [Google Scholar] [CrossRef]
- Song, X.; Wei, Y.; Xiao, D.; Gong, K.; Sun, P.; Ren, Y.; Yuan, J.; Wu, T.; Yang, Q.; Li, X.; et al. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021, 186, 388–406. [Google Scholar] [CrossRef]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW Acts as a Bifunctional Transcription Factor in Flower Development. Front. Plant Sci. 2018, 9, 835. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wang, D.; Peng, Y.; Wang, W.; Wang, Q.; Xu, Y.; Li, T.; Zhang, K.; Li, J.; Xu, X. Genome-wide analysis of the YABBY transcription factor family in rapeseed (Brassica napus L.). Genes 2021, 12, 981. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 2008, 20, 2559–2570. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
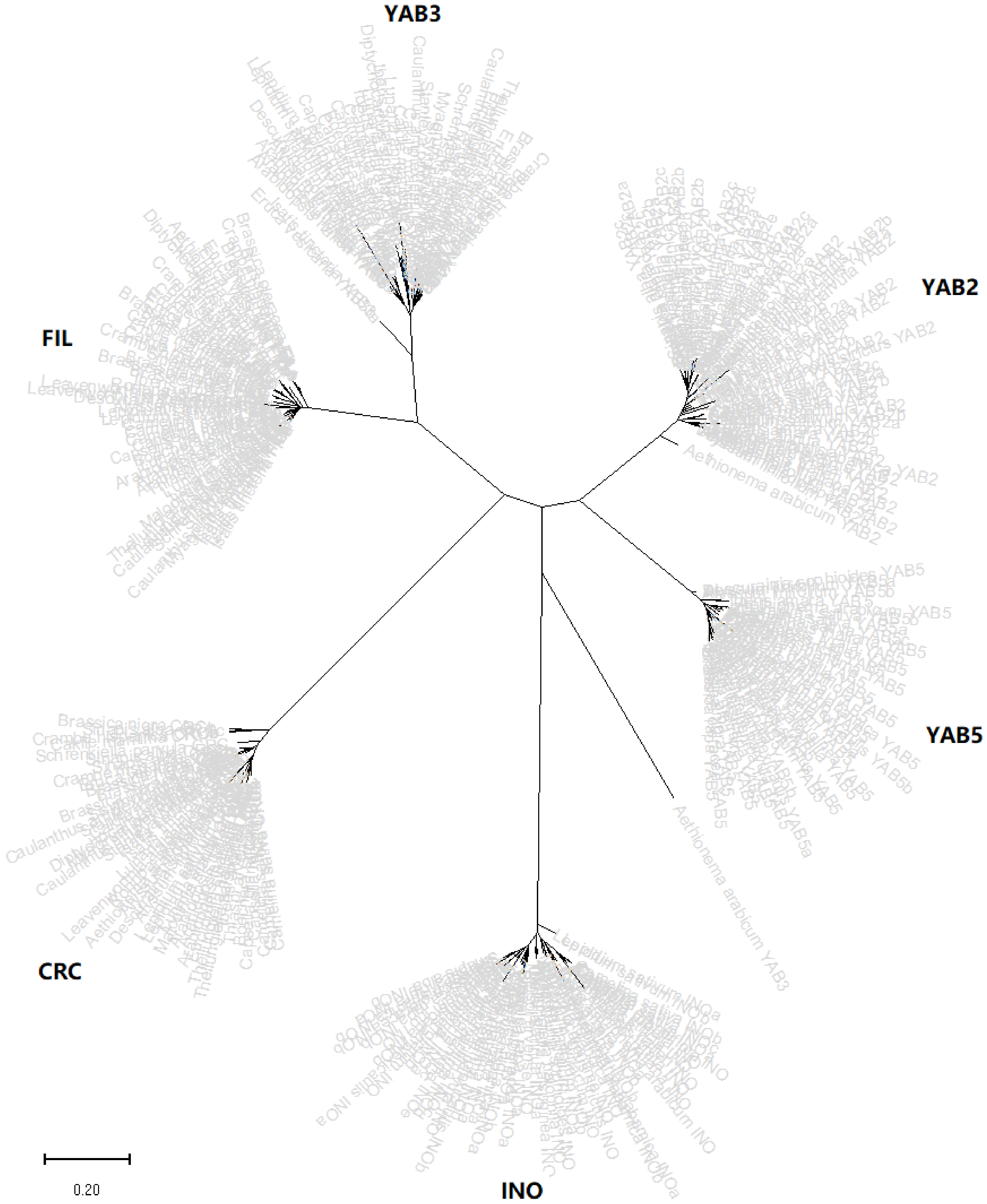
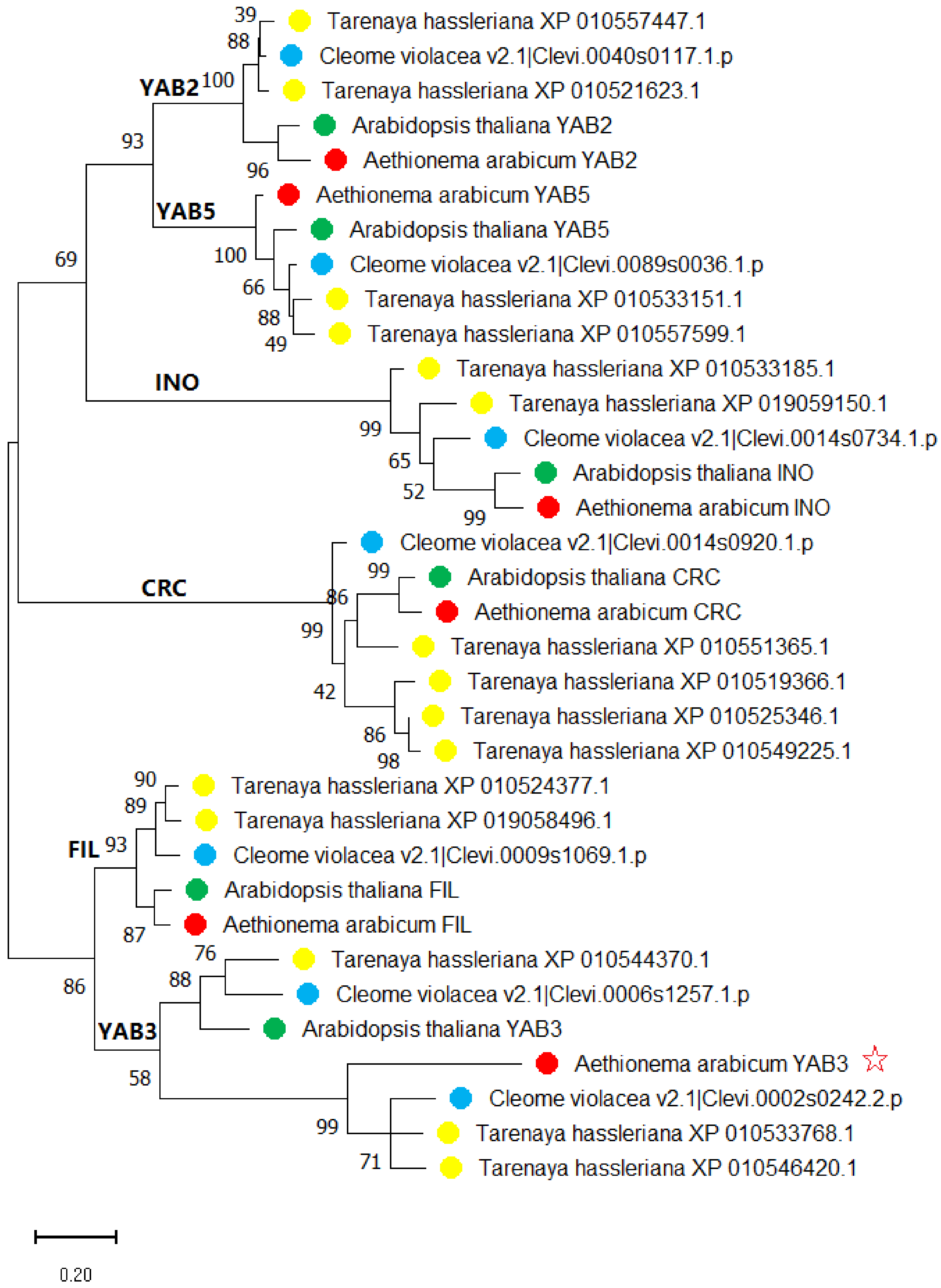

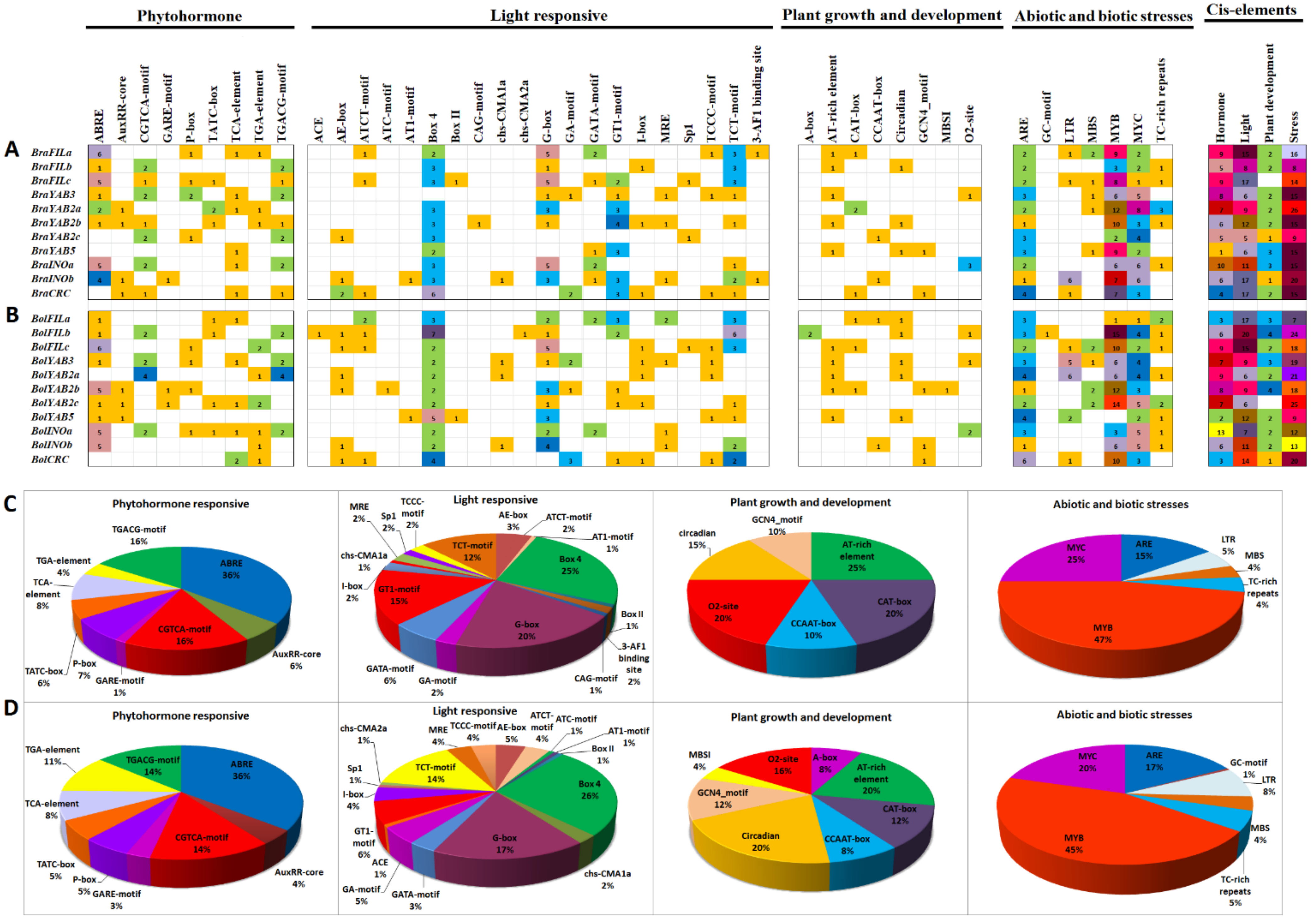
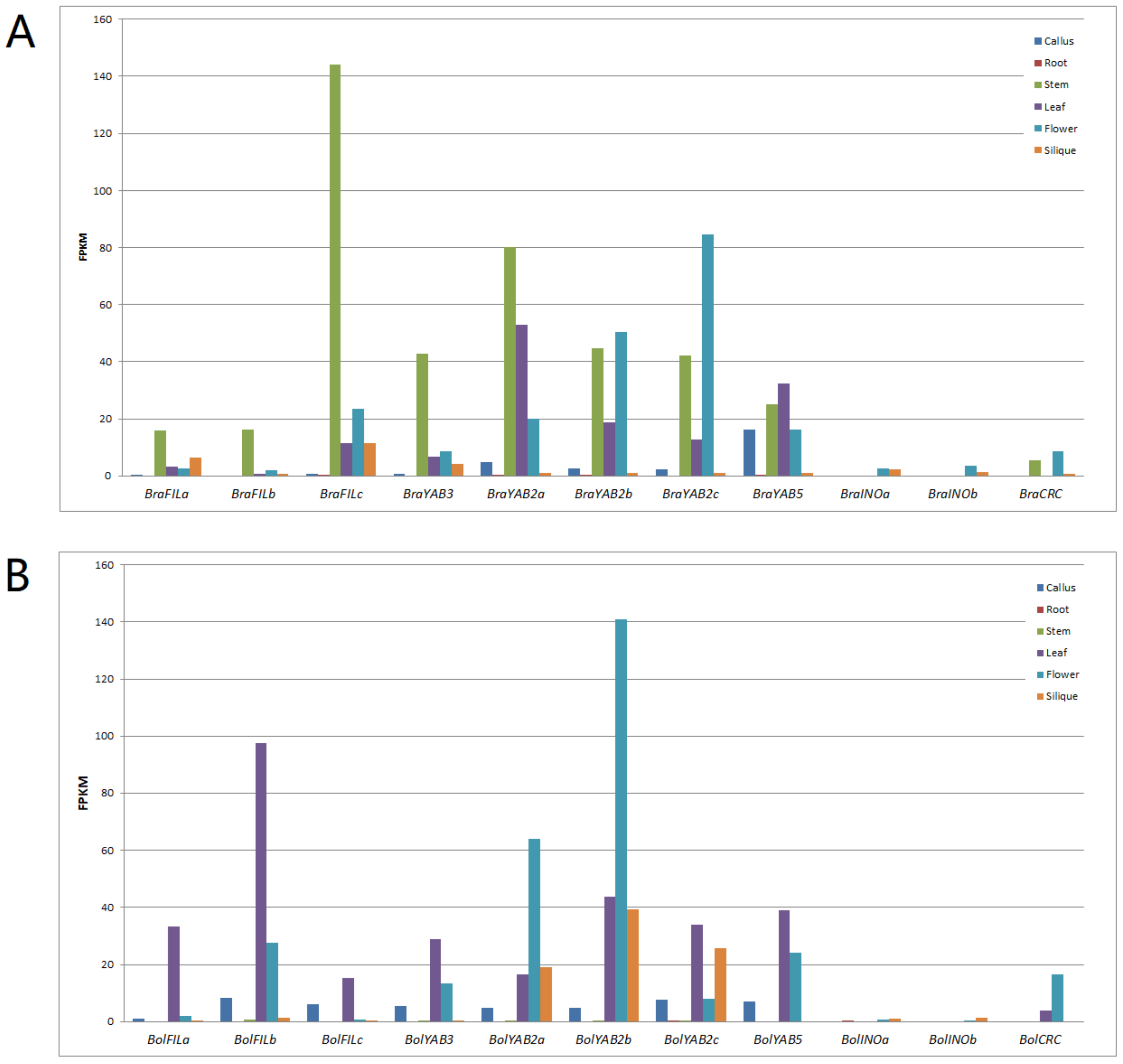
| Species | Ligneage | Clade | FIL | YAB2 | YAB3 | YAB5 | INO | CRC | Total |
|---|---|---|---|---|---|---|---|---|---|
| Aethionema arabicum | - | F | 1 | 1 | 1 a | 1 | 1 | 1 | 6 |
| Alyssum linifolium | I | D | 1 | 2 | 1 | 2 | 2 | 2 | 10 |
| Arabidopsis halleri | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Arabidopsis lyrata | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Boechera stricta | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Arabidopsis thaliana | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Brassica rapa | II | B | 3 | 3 | 1 | 1 | 2 | 1 | 11 |
| Brassica nigra | II | B | 3 | 3 | 1 | 1 | 2 | 2 | 12 |
| Brassica oleracea | II | B | 3 | 3 | 1 | 1 | 2 | 1 | 11 |
| Brassica juncea | II | B | 7 | 7 | 2 | 2 | 4 | 3 | 25 |
| Brassica napus | II | B | 6 | 6 | 2 | 2 | 4 | 2 | 22 |
| Brassica carinata | II | B | 4 | 4 | 2 | 1 | 4 | 1 | 16 |
| Cakile maritima | II | B | 3 | 3 | 2 | 1 | 5 | 2 | 16 |
| Camelina sativa | I | A | 3 | 3 | 3 | 3 | 3 | 3 | 18 |
| Capsella grandiflora | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Capsella rubella | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Caulanthus amplexicaulis | II | B | 2 | 2 | 2 | 2 | 2 | 2 | 12 |
| Crambe hispanica | II | B | 3 | 3 | 1 | 1 | 2 | 2 | 12 |
| Descurainia sophioides | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Diptychocarpus strictus | III | E | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Eruca vesicaria | II | B | 5 | 6 | 2 | 1 | 2 | 1 | 17 |
| Euclidium syriacum | III | E | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Iberis amara | II | C | 2 | 3 | 2 | 1 | 2 | 1 | 11 |
| Isatis tinctoria | II | B | 4 | 2 | 3 | 1 | 3 | 2 | 15 |
| Leavenworthia alabamica | I | A | 2 | 1 | 0 | 1 | 2 | 1 | 7 |
| Lepidium sativum | I | A | 2 | 2 | 2 | 0 | 2 | 2 | 10 |
| Lunaria annua | II | C | 3 | 2 | 2 | 1 | 2 | 2 | 12 |
| Malcolmia maritima | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Myagrum perfoliatum | II | B | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Rorippa islandica | I | A | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Schrenkiella parvula | II | B | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Sinapis alba | II | B | 3 | 3 | 1 | 1 | 2 | 3 | 13 |
| Sisymbrium irio | II | B | 1 | 2 b | 1 | 1 | 1 | 1 | 7 |
| Stanleya pinnata | II | B | 0 | 0 | 1 | 2 | 0 | 2 | 5 |
| Thellungiella halophila | II | B | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Thellungiella salsuginea | II | B | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Thlaspi arvense | II | B | 1 | 1 | 1 | 1 | 1 | 1 | 6 |
| Total | 77 | 77 | 49 | 43 | 65 | 53 | 364 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.-H.; Alam, I.; Yang, Y.-Q.; Yu, Y.-C.; Chi, W.-C.; Chen, S.-B.; Chalhoub, B.; Jiang, L.-X. Evolutionary Analysis of the YABBY Gene Family in Brassicaceae. Plants 2021, 10, 2700. https://doi.org/10.3390/plants10122700
Lu Y-H, Alam I, Yang Y-Q, Yu Y-C, Chi W-C, Chen S-B, Chalhoub B, Jiang L-X. Evolutionary Analysis of the YABBY Gene Family in Brassicaceae. Plants. 2021; 10(12):2700. https://doi.org/10.3390/plants10122700
Chicago/Turabian StyleLu, Yun-Hai, Intikhab Alam, Yan-Qing Yang, Ya-Cen Yu, Wen-Chao Chi, Song-Biao Chen, Boulos Chalhoub, and Li-Xi Jiang. 2021. "Evolutionary Analysis of the YABBY Gene Family in Brassicaceae" Plants 10, no. 12: 2700. https://doi.org/10.3390/plants10122700







