Relative Telomere Length and Telomerase Reverse Transcriptase (TERT) Expression Are Associated with Age in Almond (Prunus dulcis [Mill.] D.A.Webb)
Abstract
:1. Introduction
2. Results
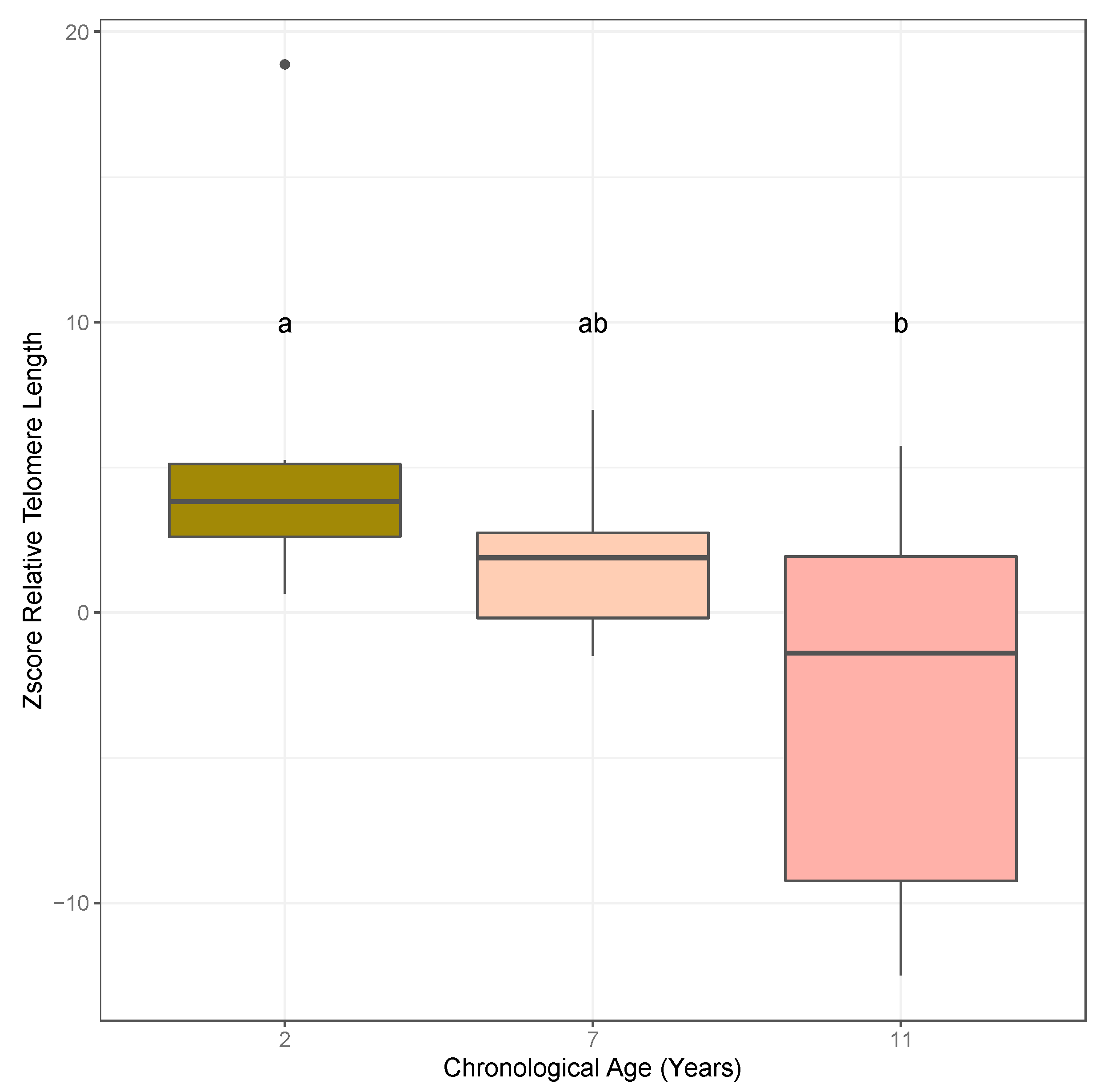
2.1. Association of Relative Telomere Length and Age in Almond
2.2. TERT Gene Expression Patterns Associated with Age in Almond
3. Discussion
3.1. Quantitative PCR Approaches Suggest an Association between Relative Telomere Length and Age in Almond Leaf and Bud Tissues
3.2. TERT Expression Measured by qRT-PCR Is Putatively Associated with Age in Almond Accessions
4. Materials and Methods
4.1. Plant Material
4.2. DNA and RNA Extraction
4.3. Monochrome Multiplex Quantitative PCR (MMQPCR) to Measure Relative Telomere Lengths
4.4. cDNA Synthesis and Quantitative Reverse Transcriptase PCR (qRT-PCR) to Measure Relative Expression of TERT
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirkwood, T.B.L. Understanding the odd science of aging. Cell 2005, 120, 437–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, J.L.; Newman, A.B. Telomere length in epidemiology: A biomarker of aging, age-related disease, both, or neither? Epidemiol. Rev. 2013, 35, 112–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munné-Bosch, S. Aging in perennials. CRC Crit. Rev. Plant Sci. 2007, 26, 123–138. [Google Scholar] [CrossRef]
- Brutovská, E.; Sámelová, A.; Dušička, J.; Mičieta, K. Ageing of trees: Application of general ageing theories. Ageing Res. Rev. 2013, 12, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Poethig, R.S. Phase change and the regulation of developmental timing in plants. Science 2003, 301, 334–336. [Google Scholar] [CrossRef]
- Almond Board of California. Almond Almanac; Almond Board of California: Modesto, CA, USA, 2019. [Google Scholar]
- Micke, W.C. Almond Production Manual; UCANR Publications: Davis, CA, USA, 1996; ISBN 978-187-990-622-8. [Google Scholar]
- Wickson, E.J. The California Fruits and How to Grow Them; Pacific Rural: San Francisco, CA, USA, 1914; ISBN 978-198-522-645-6. [Google Scholar]
- Kester, D.E.; Jones, R.W. Non-infectious bud-failure from breeding programs of almond (Prunus amygdalus Batsch). J. Am. Soc. Hort. Sci. 1970, 95, 492–496. [Google Scholar]
- Gradziel, T.M.; Thorpe, M.A.; Fresnedo-Ramírez, J.; Ragas, R.; Lampinen, B.; Adaskaveg, J.; Connell, J.; Schrader, P.; Metcalf, S.; Duncan, R.; et al. Molecular Marker Based Diagnostics for Almond Non-Infectious Bud-Failure; Almond Board of California: Modesto, CA, USA, 2013. [Google Scholar]
- Kester, D.E.; Shackel, K.A.; Micke, W.C.; Viveros, M.; Gradziel, T.M. Noninfectious bud failure in ‘Carmel’ almond: I. Pattern of development in vegetative progeny trees. J. Am. Soc. Hort. Sci. 2004, 129, 244–249. [Google Scholar] [CrossRef]
- Bilova, T.; Paudel, G.; Shilyaev, N.; Schmidt, R.; Brauch, D.; Tarakhovskaya, E.; Milrud, S.; Smolikova, G.; Tissier, A.; Vogt, T.; et al. Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. J. Biol. Chem. 2017, 292, 15758–15776. [Google Scholar] [CrossRef] [Green Version]
- Dubrovina, A.S.; Kiselev, K.V.; Weber, A. Age-associated alterations in the somatic mutation and DNA methylation levels in plants. Plant Biol. 2016, 18, 185–196. [Google Scholar] [CrossRef]
- Marioni, R.E.; Harris, S.E.; Shah, S.; McRae, A.F.; von Zglinicki, T.; Martin-Ruiz, C.; Wray, N.R.; Visscher, P.M.; Deary, I.J. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int. J. Epidemiol. 2016, 45, 424–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runov, A.L.; Vonsky, M.S.; Mikhelson, V.M. DNA methylation level and telomere length as a basis for modeling of the biological aging clock. Cell Tissue Biol. 2015, 9, 261–264. [Google Scholar] [CrossRef]
- Watson, J.M.; Riha, K. Telomeres, aging, and plants: From weeds to Methuselah—A mini-review. Gerontology 2011, 57, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.D.L.; Beilstein, M.A.; Shippen, D.E. Plant telomeres and telomerase. In Molecular Biology; Howell, S.H., Ed.; Springer: New York, NY, USA, 2014; pp. 25–49. ISBN 978-1-4614-7570-5. [Google Scholar]
- Aviv, A.; Shay, J.W. Reflections on telomere dynamics and ageing-related diseases in humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, M.S.; McKnight, T.D.; Shippen, D.E. Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. USA 1996, 93, 14422–14427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Torre-Espinosa, Z.Y.; Barredo-Pool, F.; Castaño de la Serna, E.; Sánchez-Teyer, L.F. Active telomerase during leaf growth and increase of age in plants from Agave tequilana var. Azul. Physiol. Mol. Biol. Plants 2020, 26, 639–647. [Google Scholar] [CrossRef]
- Boccardi, V.; Paolisso, G. Telomerase activation: A potential key modulator for human healthspan and longevity. Ageing Res. Rev. 2014, 15, 1–5. [Google Scholar] [CrossRef]
- Anchelin, M.; Murcia, L.; Alcaraz-Pérez, F.; García-Navarro, E.M.; Cayuela, M.L. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Fossel, M. Telomerase and the aging cell: Implications for human health. JAMA J. Am. Med. Assoc. 1998, 279, 1732–1735. [Google Scholar] [CrossRef]
- Oguchi, K.; Liu, H.; Tamura, K.; Takahashi, H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 1999, 457, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Procházková Schrumpfová, P.; Fojtová, M.; Fajkus, J. Telomeres in plants and humans: Not so different, not so similar. Cells 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweetlove, L.; Gutierrez, C. The journey to the end of the chromosome: Delivering active telomerase to telomeres in plants. Plant J. 2019, 98, 193–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurečková, J.F.; Sýkorová, E.; Hafidh, S.; Honys, D.; Fajkus, J.; Fojtová, M. Tissue-specific expression of telomerase reverse transcriptase gene variants in Nicotiana tabacum. Planta 2017, 245, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, L.; Arakelyan, A. Computel: Computation of mean telomere length from whole-genome next-generation sequencing data. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montpetit, A.J.; Alhareeri, A.A.; Montpetit, M.; Starkweather, A.R.; Elmore, L.W.; Filler, K.; Mohanraj, L.; Burton, C.W.; Menzies, V.S.; Lyon, D.E.; et al. Telomere length: A review of methods for measurement. Nurs. Res. 2014, 63, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Cawthon, R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [Green Version]
- Flanary, B.E.; Kletetschka, G. Analysis of telomere length and telomerase activity in tree species of various life-spans, and with age in the bristlecone pine Pinus longaeva. Biogerontology 2005, 6, 101–111. [Google Scholar] [CrossRef]
- Moriguchi, R.; Kato, K.; Kanahama, K.; Kanayama, Y.; Kikuchi, H. Analysis of telomere lengths in apple and cherry trees. Acta Hort. 2007, 389–395. [Google Scholar] [CrossRef]
- Liang, J.; Jiang, C.; Peng, H.; Shi, Q.; Guo, X.; Yuan, Y.; Huang, L. Analysis of the age of Panax ginseng based on telomere length and telomerase activity. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Liu, D.; Qiao, N.; Song, H.; Hua, X.; Du, J.; Lu, H.; Li, F. Comparative analysis of telomeric restriction fragment lengths in different tissues of Ginkgo biloba trees of different age. J. Plant Res. 2007, 120, 523–528. [Google Scholar] [CrossRef]
- Hemann, M.T.; Strong, M.A.; Hao, L.-Y.; Greider, C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Zachová, D.; Fojtová, M.; Dvořáčková, M.; Mozgová, I.; Lermontova, I.; Peška, V.; Schubert, I.; Fajkus, J.; Sýkorová, E. Structure-function relationships during transgenic telomerase expression in Arabidopsis. Physiol. Plant. 2013, 149, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Zangi, M.; Bagherieh Najjar, M.B.; Golalipour, M.; Aghdasi, M. met1 DNA methyltransferase controls TERT gene expression: A new insight to the role of telomerase in development. Cell J. 2019, 22. [Google Scholar] [CrossRef]
- Fitzgerald, M.S.; Riha, K.; Gao, F.; Ren, S.; McKnight, T.D.; Shippen, D.E. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 1999, 96, 14813–14818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronen, T.; Ryynänen, L. Variation in telomeric repeats of Scots pine (Pinus sylvestris L.). Tree Genet. Genomes 2012, 8, 267–275. [Google Scholar] [CrossRef]
- Kilian, A.; Stiff, C.; Kleinhofs, A. Barley telomeres shorten during differentiation but grow in callus culture. Proc. Natl. Acad. Sci. USA 1995, 92, 9555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aronen, T.; Ryynänen, L. Silver birch telomeres shorten in tissue culture. Tree Genet. Genomes 2014, 10, 67–74. [Google Scholar] [CrossRef]
- Bonga, J.M. Vegetative propagation in relation to juvenility, maturity, and rejuvenation. In Tissue Culture in Forestry; Bonga, J.M., Durzan, D.J., Eds.; Springer: Dordrecht, The Netherlands, 1982; pp. 387–412. ISBN 978-94-017-3538-4. [Google Scholar]
- Gradziel, T.; Lampinen, B.; Preece, J.E. Propagation from basal epicormic meristems remediates an aging-related disorder in almond clones. Sci. Hort. 2019, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- Kilian, A.; Heller, K.; Kleinhofs, A. Development patterns of telomerase activity in barley and maize. Plant Mol. Biol. 1998, 37, 621–628. [Google Scholar] [CrossRef]
- Song, H.; Liu, D.; Li, F.; Lu, H. Season- and age-associated telomerase activity in Ginkgo biloba L. Mol. Biol. Rep. 2011, 38, 1799–1805. [Google Scholar] [CrossRef]
- Vega-Vaquero, A.; Bonora, G.; Morselli, M.; Vaquero-Sedas, M.I.; Rubbi, L.; Pellegrini, M.; Vega-Palas, M.A. Novel features of telomere biology revealed by the absence of telomeric DNA methylation. Genome Res. 2016, 26, 1047–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.K.; Cho, M.H. Epigenetic aspects of telomeric chromatin in Arabidopsis thaliana. BMB Rep. 2019, 52, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Ogrocká, A.; Polanská, P.; Majerová, E.; Janeba, Z.; Fajkus, J.; Fojtová, M. Compromised telomere maintenance in hypomethylated Arabidopsis thaliana plants. Nucleic Acids Res. 2014, 42, 2919–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilanova, S.; Alonso, D.; Gramazio, P.; Plazas, M.; García-Fortea, E.; Ferrante, P.; Schmidt, M.; Díez, M.J.; Usadel, B.; Giuliano, G.; et al. SILEX: A fast and inexpensive high-quality DNA extraction method suitable for multiple sequencing platforms and recalcitrant plant species. Plant Methods 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Vaquero-Sedas, M.I.; Vega-Palas, M.A. Determination of Arabidopsis thaliana telomere length by PCR. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hao, R.; Pan, H.; Cheng, T.; Zhang, Q. Selection of suitable reference genes for quantitative real-time polymerase chain reaction in Prunus mume during flowering stages and under different abiotic stress conditions. J. Am. Soc. Hort. Sci. 2014, 139, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Bastias, A.; Oviedo, K.; Almada, R.; Correa, F.; Sagredo, B. Identifying and validating housekeeping hybrid Prunus spp. genes for root gene-expression studies. PLoS ONE 2020, 15. [Google Scholar] [CrossRef] [Green Version]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [Green Version]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Martí, Á.F.; Frias, L.; et al. Transposons played a major role in the diversification between the closely related almond and peach genomes: Results from the almond genome sequence. Plant J. 2020, 101, 455–472. [Google Scholar] [CrossRef] [Green Version]
- Verhulst, S. Improving comparability between qPCR-based telomere studies. Mol. Eco. Res. 2020, 20, 11–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohio Supercomputer Center: Columbus, OH, USA. 1987. Available online: https://www.osc.edu/resources/getting_started/citation (accessed on 20 November 2020).



| Sampling Year | Age (Years) | Leaf Sample Size | Bud Sample Size |
|---|---|---|---|
| 2018 | 1 | 6 | N/A |
| 2018 | 5 | 6 | N/A |
| 2018 | 9 | 6 | N/A |
| 2018 | 14 | 6 | N/A |
| 2019 | 2 | 4 | 6 |
| 2019 | 7 | 4 | 7 |
| 2019 | 11 | 4 | 5 |
| Oligo Name | Oligo Sequence (5′–3′) |
|---|---|
| PP2A Forward | CGGCGGCGGGCGGCGCGGGCAGGATAGACATTGGAGGGTTCGGCTCGCAA |
| PP2A Reverse | CGGCGGCGGGCGGCGCGGGACCACTGCATGCAAAGGGACCCAAGCTTAT |
| Telomere Forward | CCCCGGTTTTGGGTTTTGGGTTTTGGGTTTTGGGT |
| Telomere Reverse | GGGGCCCTAATCCCTAATCCCTAATCCCTAATCCCT |
| TERT Forward | GCATCAGAGAAGGGTCAGATT |
| TERT Reverse | CTCTGGCTCCTTGAATCGTATAG |
| RPII Forward | TGAAGCATACACCTATGATGATGAAG |
| RPII Reverse | CTTTGACAGCACCAGTAGATTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D'Amico-Willman, K.M.; Anderson, E.S.; Gradziel, T.M.; Fresnedo-Ramírez, J. Relative Telomere Length and Telomerase Reverse Transcriptase (TERT) Expression Are Associated with Age in Almond (Prunus dulcis [Mill.] D.A.Webb). Plants 2021, 10, 189. https://doi.org/10.3390/plants10020189
D'Amico-Willman KM, Anderson ES, Gradziel TM, Fresnedo-Ramírez J. Relative Telomere Length and Telomerase Reverse Transcriptase (TERT) Expression Are Associated with Age in Almond (Prunus dulcis [Mill.] D.A.Webb). Plants. 2021; 10(2):189. https://doi.org/10.3390/plants10020189
Chicago/Turabian StyleD'Amico-Willman, Katherine M., Elizabeth S. Anderson, Thomas M. Gradziel, and Jonathan Fresnedo-Ramírez. 2021. "Relative Telomere Length and Telomerase Reverse Transcriptase (TERT) Expression Are Associated with Age in Almond (Prunus dulcis [Mill.] D.A.Webb)" Plants 10, no. 2: 189. https://doi.org/10.3390/plants10020189
APA StyleD'Amico-Willman, K. M., Anderson, E. S., Gradziel, T. M., & Fresnedo-Ramírez, J. (2021). Relative Telomere Length and Telomerase Reverse Transcriptase (TERT) Expression Are Associated with Age in Almond (Prunus dulcis [Mill.] D.A.Webb). Plants, 10(2), 189. https://doi.org/10.3390/plants10020189





