An Efficient Method for the Genetic Transformation of Acmella oleracea L. (Spilanthes acmella Linn.) with Agrobacterium tumefaciens
Abstract
1. Introduction
2. Results and Discussion
2.1. Explant Effect on the Regeneration by Organogenesis in A. oleracea
2.2. Agrobacterium-Mediated Transformation
2.2.1. Determination of a Kanamycin Dose–Response Curve for the Selection of Acmella pBI121 Transformants
2.2.2. Nodal Explant Infection
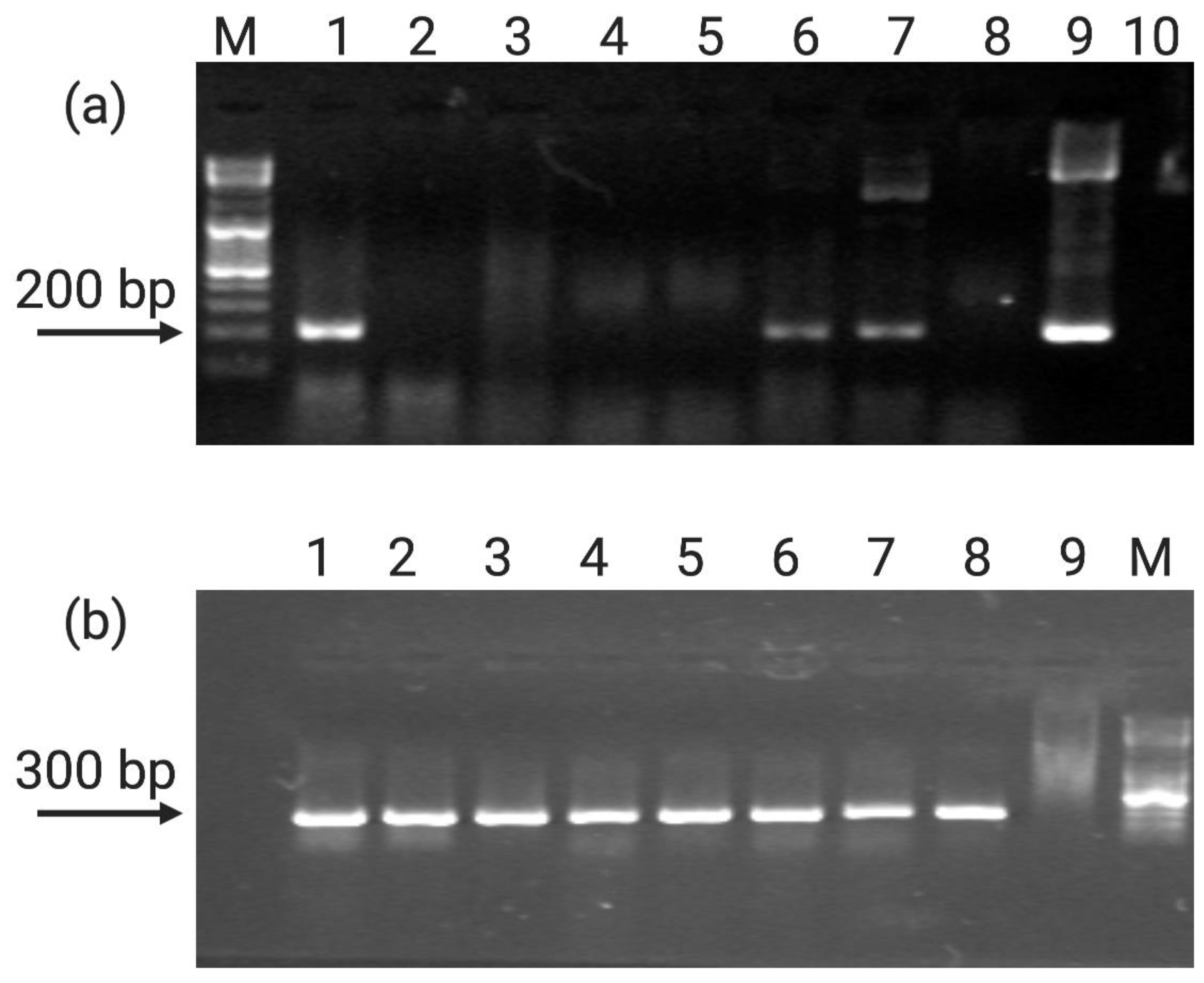
2.3. Molecular Analysis of Putative Transformants
2.3.1. Analysis of the Presence of Transgenes by Direct PCR
2.3.2. GUS Gene Expression Analysis
3. Materials and Methods
3.1. Plant Material and In Vitro Culture Establishment
3.2. Regeneration by Organogenesis of A. oleracea
3.3. Agrobacterium-Mediated Transformation of A. oleracea
3.3.1. Determination of Kanamycin Concentration for the Selection of Transformants
3.3.2. Leaf Disc Transformation and Molecular Analysis of Transformants
3.3.3. GUS Assay with X-Gluc and GUS Expression Analysis with RT-PCR
3.4. Statistical Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jansen, R.K. The Systematics of Acmella (Asteraceae-Heliantheae). Syst. Bot. Monogr. 1985, 8, 1. [Google Scholar] [CrossRef]
- Singh, M.; Chaturvedi, R. Screening and quantification of an antiseptic alkylamide, spilanthol from in vitro cell and tissue cultures of Spilanthes acmella Murr. Ind. Crops Prod. 2012, 36, 321–328. [Google Scholar] [CrossRef]
- Singh, M.; Chaturvedi, R. Evaluation of nutrient uptake and physical parameters on cell biomass growth and production of spilanthol in suspension cultures of Spilanthes acmella Murr. Bioprocess Biosyst. Eng. 2012, 35, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Abeysiri, G.R.P.I.; Dharmadasa, R.M.; Abeysinghe, D.C.; Samarasinghe, K. Screening of phytochemical, physico-chemical and bioactivity of different parts of Acmella oleraceae Murr. (Asteraceae), a natural remedy for toothache. Ind. Crops Prod. 2013, 50, 852–856. [Google Scholar] [CrossRef]
- Martins, C.P.S.; Melo, M.T.P.; Honório, I.C.G.; D’Ávila, V.A.; Carvalho, W.G. Caracterização morfológica e agronômica de acessos de jambu (Spilanthes oleracea L.) nas condições do Norte de Minas Gerais. Rev. Bras. Plantas Med. 2012, 14, 410–413. [Google Scholar] [CrossRef]
- Tiwari, K.L.; Jadhav, S.K.; Joshi, V. An updated review on medicinal herb genus Spilanthes. J. Chin. Integr. Med. 2011, 9, 1170–1178. [Google Scholar] [CrossRef]
- Paulraj, J.; Govindarajan, R.; Palpu, P. The genus Spilanthes ethnopharmacology, phytochemistry, and pharmacological properties: A review. Adv. Pharmacol. Sci. 2013, 2013, 510298. [Google Scholar]
- Prachayasittikul, V.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. High therapeutic potential of Spilanthes acmella: A review. EXCLI J. 2013, 12, 291–312. [Google Scholar]
- Lalchhandama, K.; Lalthanpuii, P.B.; Zokimi, Z. The toothache plant (Acmella oleracea) exhibits anthelmintic activity on both parasitic tapeworms and roundworms. Pharmacog. Mag. 2020, 16, 193. [Google Scholar] [CrossRef]
- Uthpala, T.G.G.; Navaratne, S.B. Acmella oleracea plant; Identification, Applications and Use as an Emerging Food Source—Review. Food Rev. Int. 2020, 1–16. [Google Scholar] [CrossRef]
- Lalthanpuii, P.B.; Lalchhandama, K. Chemical composition and broad-spectrum anthelmintic activity of a cultivar of toothache plant, Acmella oleracea, from Mizoram, India. Pharm. Biol. 2020, 58, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Dallazen, J.L.; Maria-Ferreira, D.; da Luz, B.B.; Nascimento, A.M.; Cipriani, T.R.; de Souza, L.M.; Felipe, L.P.G.; Silva, B.J.G.; Nassini, R.; de Paula Werner, M.F. Pharmacological potential of alkylamides from Acmella oleracea flowers and synthetic isobutylalkyl amide to treat inflammatory pain. Inflammopharmacology 2020, 28, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Lalthanpuii, P.B.; Lalchhandama, K. Intestinal cestodes of chicken are effectively killed by quinoline-rich extract of Spilanthes acmella. Vet. World 2020, 13, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Dallazen, J.L.; Maria-Ferreira, D.; da Luz, B.B.; Nascimento, A.M.; Cipriani, T.R.; de Souza, L.M.; Glugoski, L.P.; Silva, B.J.G.; Geppetti, P.; de Paula Werner, M.F. Distinct mechanisms underlying local antinociceptive and pronociceptive effects of natural alkylamides from Acmella oleracea compared to synthetic isobutylalkyl amide. Fitoterapia 2018, 131, 225–235. [Google Scholar] [CrossRef]
- Dias, A.M.A.; Santos, P.; Seabra, I.J.; Júnior, R.N.C.; Braga, M.E.M.; de Sousa, H.C. Spilanthol from Spilanthes acmella flowers, leaves and stems obtained by selective supercritical carbon dioxide extraction. J. Supercrit. Fluids 2012, 61, 62–70. [Google Scholar] [CrossRef]
- Simas, N.K.; da Costa Lima Dellamora, E.; Schripsema, J.; Lage, C.L.S.; de Oliveira Filho, A.M.; Wessjohann, L.; Porzel, A.; Kuster, R.M. Acetylenic 2-phenylethylamides and new isobutylamides from Acmella oleracea (L.) R. K. Jansen, a Brazilian spice with larvicidal activity on Aedes aegypti. Phytochem. Lett. 2013, 6, 67–72. [Google Scholar] [CrossRef]
- Dubey, S.; Maity, S.; Singh, M.; Saraf, S.A.; Saha, S. Phytochemistry, pharmacology and toxicology of Spilanthes acmella: A Review. Adv. Pharmacol. Sci. 2013, 2013, 423750. [Google Scholar]
- Joshi, V.; Sharma, G.D.; Jadhav, S.K. Alkamides: Multifunctional bioactive agents in Spilanthes spp. J. Sci. Res. 2020, 64, 198–206. [Google Scholar] [CrossRef]
- Barbosa, A.F.; de Carvalho, M.G.; Smith, R.E.; Sabaa-Srur, A.U.O. Spilanthol: Occurrence, extraction, chemistry and biological activities. Rev. Bras. Farmacogn. 2016, 26, 128–133. [Google Scholar] [CrossRef]
- Nascimento, L.E.S.; Arriola, N.D.A.; da Silva, L.A.L.; Faqueti, L.G.; Sandjo, L.P.; de Araújo, C.E.S.; Biavatti, M.W.; Barcelos-Oliveira, J.L.; de Mello Castanho Amboni, R.D. Phytochemical profile of different anatomical parts of jambu (Acmella oleracea (L.) R.K. Jansen): A comparison between hydroponic and conventional cultivation using PCA and cluster analysis. Food Chem. 2020, 332, 127393. [Google Scholar] [CrossRef]
- Sut, S.; Ferrarese, I.; Shrestha, S.S.; Kumar, G.; Slaviero, A.; Sello, S.; Altissimo, A.; Pagni, L.; Gattesco, F.; Dall’Acqua, S. Comparison of biostimulant treatments in Acmella oleracea cultivation for alkylamides production. Plants 2020, 9, 818. [Google Scholar] [CrossRef] [PubMed]
- Rios-Chavez, P.; Ramirez-Chavez, E.; Armenta-Salinas, C.; Molina-Torres, J. Acmella radicans var. radicans: In vitro culture establishment and alkamide content. In Vitro Cell. Dev. Biol. Plant 2003, 39, 37–41. [Google Scholar] [CrossRef]
- Saritha, K.V.; Naidu, C.V. Direct shoot regeneration from leaf explants of Spilanthes acmella. Biol. Plant. 2008, 52, 334–338. [Google Scholar] [CrossRef]
- Deka, P.; Kalita, M.C. In vitro Clonal propagation and organogenesis in Spilanthes acmella (L) Murray: A herbal pesticidal plant of North-East India. J. Plant Biochem. Biotech. 2005, 14, 69–71. [Google Scholar] [CrossRef]
- Pandey, V.; Agrawal, V. Efficient micropropagation protocol of Spilanthes acmella L. possessing strong antimalarial activity. In Vitro Cell. Dev. Biol. Plant 2009, 45, 491–499. [Google Scholar] [CrossRef]
- Lima, N.K.; Da Silva, E.S.; Da Cruz, R.M.S.; Monteiro, P.H.R.; Da Silva, G.J. Plant growth regulators in the in vitro cultivation of Acmella oleracea (L.). J. Agric. Stud. 2020, 8, 774. [Google Scholar] [CrossRef]
- Almeida, S.P.; Souza, J.M.M.; Amorim, A.M.T.; de Gusmão, S.A.L.; Souza, R.O.; Santos, A.S. In vitro culture of jambu with different growth regulators. Hortic. Bras. 2020, 38, 134–138. [Google Scholar] [CrossRef]
- Singh, R.S.; Chattopadhyay, T.; Thakur, D.; Kumar, N.; Kumar, T.; Singh, P.K. Hairy root culture for in vitro production of secondary metabolites: A promising biotechnological approach. In Biotechnological Approaches for Medicinal and Aromatic Plants. Conservation, Genetic Improvement and Utilization; Kumar, N., Ed.; Springer: Singapore, 2018; pp. 235–250. [Google Scholar]
- Li, C.; Wang, M. Application of hairy root culture for bioactive compounds production in medicinal plants. Curr. Pharm. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Shi, T.; Shi, J.; Xia, Z.; Lu, B.; Shi, S.; Fu, G. Precise control of variable-height laser metal deposition using a height memory strategy. J. Manuf. Processes 2020, 57, 222–232. [Google Scholar] [CrossRef]
- Wilson, S.A.; Roberts, S.C. Metabolic engineering approaches for production of biochemicals in food and medicinal plants. Curr. Opin. Biotechnol. 2014, 26, 174–182. [Google Scholar] [CrossRef]
- Alok, A.; Jain, P.; Kumar, J.; Yajnik, K.; Bhalothia, P. Genome engineering in medicinally important plants using CRISPR/Cas9 tool. In Genome Engineering via CRISPR-Cas9 System, 1st ed.; Singh, V., Dhar, P.K., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 155–161. [Google Scholar]
- Khan, S.; Rahman, L.u. Pathway Modulation of medicinal and aromatic plants through metabolic engineering using Agrobacterium tumefaciens. In Transgenesis and Secondary Metabolism; Reference Series in Phytochemistry; Jha, S., Ed.; Springer: Cham, Switzerland, 2016; pp. 1–32. [Google Scholar]
- Niazian, M. Application of genetics and biotechnology for improving medicinal plants. Planta 2019, 249, 953–973. [Google Scholar] [CrossRef] [PubMed]
- Saritha, K.V.; Prakash, E.; Ramamurthy, N.; Naidu, C.V. Micropropagation of Spilanthes acmella Murr. Biol. Plant. 2002, 45, 581–584. [Google Scholar] [CrossRef]
- Singh, M.; Chaturvedi, R. Optimization of Sphilanthes acmella L. cultivation by in vitro nodal segment culture. Acta Hortic. 2010, 109–114. [Google Scholar] [CrossRef]
- Singh, S.K.; Rai, M.K.; Asthana, P.; Pandey, S.; Jaiswal, V.S.; Jaiswal, U. Plant regeneration from alginate-encapsulated shoot tips of Spilanthes acmella (L.) Murr., a medicinally important and herbal pesticidal plant species. Acta Physiol. Plant. 2009, 31, 649–653. [Google Scholar] [CrossRef]
- Singh, S.K.; Rai, M.K.; Asthana, P.; Sahoo, L. An improved micropropagation of Spilanthes acmella L. through transverse thin cell layer culture. Acta Physiol. Plant. 2009, 31, 693–698. [Google Scholar] [CrossRef]
- Matzk, A.; Mantell, S.; Schiemann, J. Localization of persisting agrobacteria in transgenic tobacco plants. MPMI-Mol. Plant Microbe Interact. 1996, 9, 373–381. [Google Scholar] [CrossRef]
- Praveen, M.; Nanna, R.S. Influence of antibiotics on regeneration efficiency in tomato. Plant Omics 2009, 2, 135. [Google Scholar]
- Retheesh, S.T.; Bhat, A.I. Genetic transformation and regeneration of transgenic plants from protocorm-like bodies of vanilla (Vanilla planifolia Andrews) using Agrobacterium tumefaciens. J. Plant Biochem. Biotechnol. 2011, 20, 262. [Google Scholar] [CrossRef]
- Priya, A.M.; Pandian, S.K.; Manikandan, R. The effect of different antibiotics on the elimination of Agrobacterium and high frequency Agrobacterium-mediated transformation of indica rice (Oryza sativa L.). Czech J. Genet. Plant Breed. 2012, 48, 120–130. [Google Scholar] [CrossRef]
- Bettini, P.P.; Chiarugi, P.; Buiatti, M. An in vitro molecular study of the Nicotiana tabacum L. genome in the presence or absence of the herbicide atrazine. Theor. Appl. Genet. 1998, 96, 242–250. [Google Scholar] [CrossRef]
- Linsmaier, E.M.; Skoog, F. Organic Growth Factor Requirements of tobacco tissue cultures. Physiol. Plant. 1965, 18, 100–127. [Google Scholar] [CrossRef]
- Ooms, G.; Hooykaas, P.J.; Van Veen, R.J.; Van Beelen, P.; Regensburg-Tuïnk, T.J.; Schilperoort, R.A. Octopine Ti-plasmid deletion mutants of Agrobacterium tumefaciens with emphasis on the right side of the T-region. Plasmid 1982, 7, 15–29. [Google Scholar] [CrossRef]
- Chen, P.Y.; Chen-Kuen, W.; Shaw-Ching, S.; Kin-Ying, T. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 2003, 11, 287–293. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST-PAlaeontological STatistics, ver. 1.89. Palaeontol. Electron. 2009, 4, 1–9. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maggini, V.; Bettini, P.; Firenzuoli, F.; Bogani, P. An Efficient Method for the Genetic Transformation of Acmella oleracea L. (Spilanthes acmella Linn.) with Agrobacterium tumefaciens. Plants 2021, 10, 198. https://doi.org/10.3390/plants10020198
Maggini V, Bettini P, Firenzuoli F, Bogani P. An Efficient Method for the Genetic Transformation of Acmella oleracea L. (Spilanthes acmella Linn.) with Agrobacterium tumefaciens. Plants. 2021; 10(2):198. https://doi.org/10.3390/plants10020198
Chicago/Turabian StyleMaggini, Valentina, Priscilla Bettini, Fabio Firenzuoli, and Patrizia Bogani. 2021. "An Efficient Method for the Genetic Transformation of Acmella oleracea L. (Spilanthes acmella Linn.) with Agrobacterium tumefaciens" Plants 10, no. 2: 198. https://doi.org/10.3390/plants10020198
APA StyleMaggini, V., Bettini, P., Firenzuoli, F., & Bogani, P. (2021). An Efficient Method for the Genetic Transformation of Acmella oleracea L. (Spilanthes acmella Linn.) with Agrobacterium tumefaciens. Plants, 10(2), 198. https://doi.org/10.3390/plants10020198




