The Ubiquitin Switch in Plant Stress Response
Abstract
1. Introduction
2. Protein Turnover by the Plant UPS
3. Crosstalk of Ub with Its Two Siblings, ATG8 and SUMO, in Protein Turnover
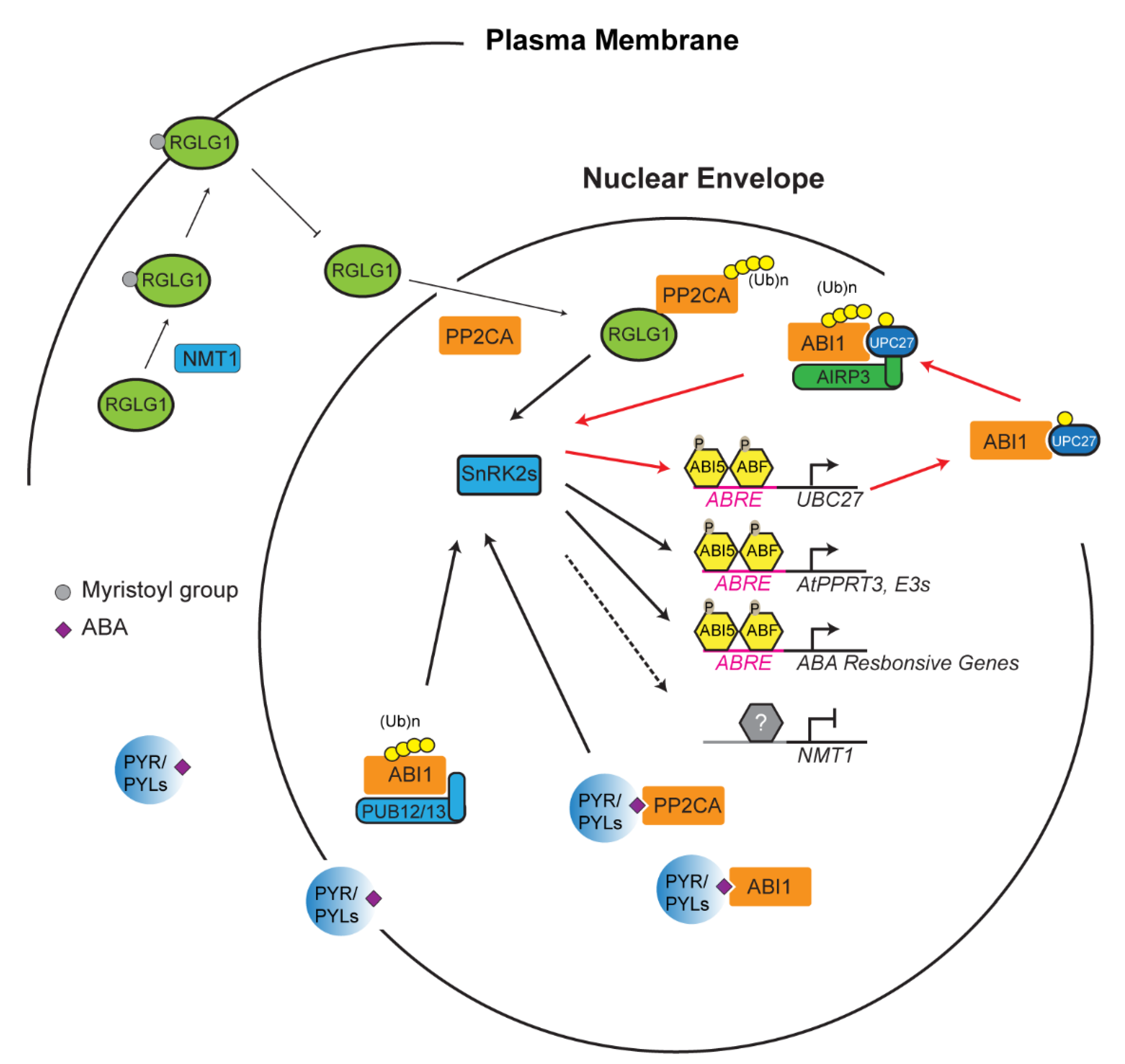
4. Reinforcement Intersection of the UPS with ABA in Abiotic Stress Response
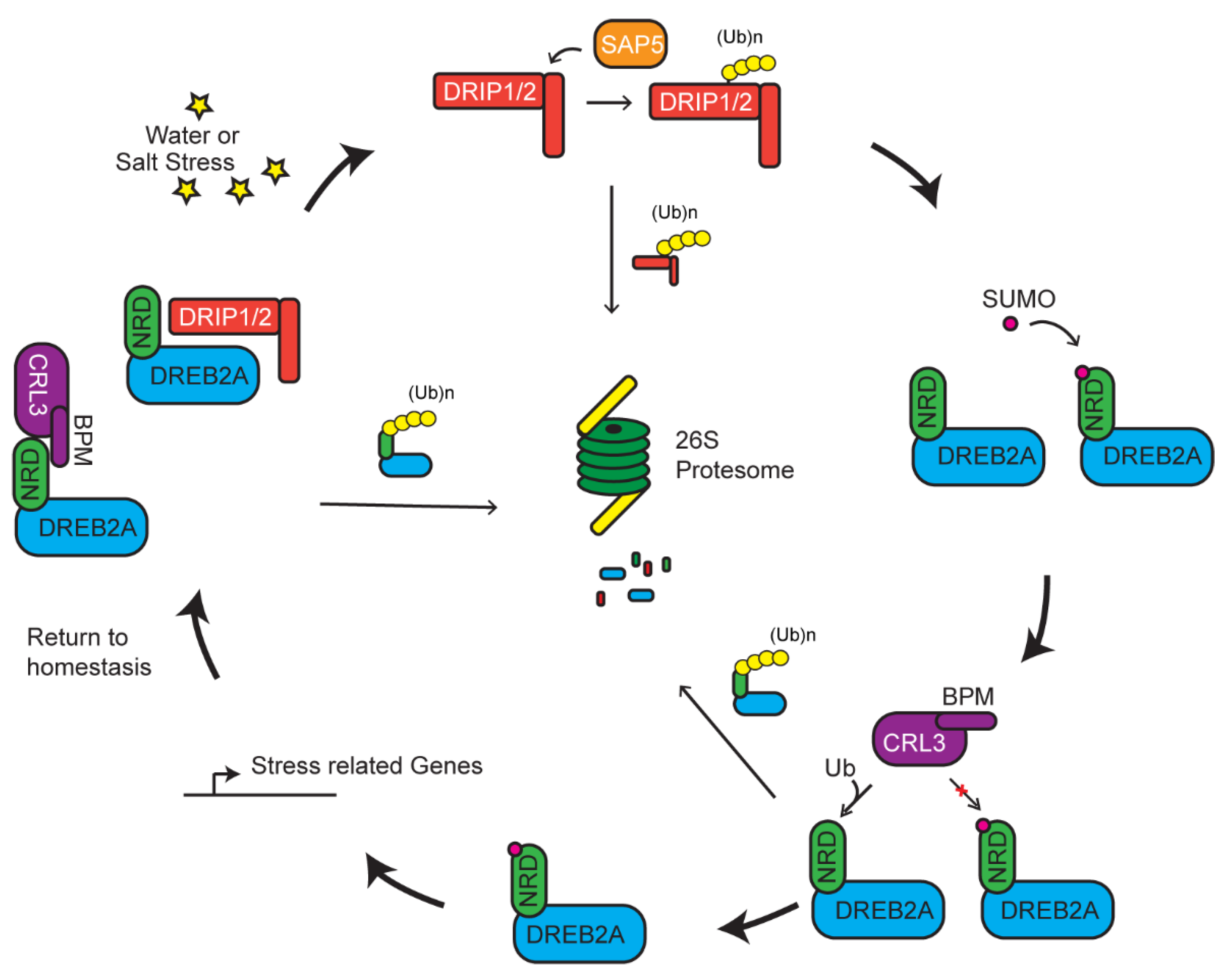
5. Multiple UPS Routes Acting Upon DREB2A-Mediated Stress Tolerance
6. Influence of the UPS in Stress Regulation through ROS Homeostasis
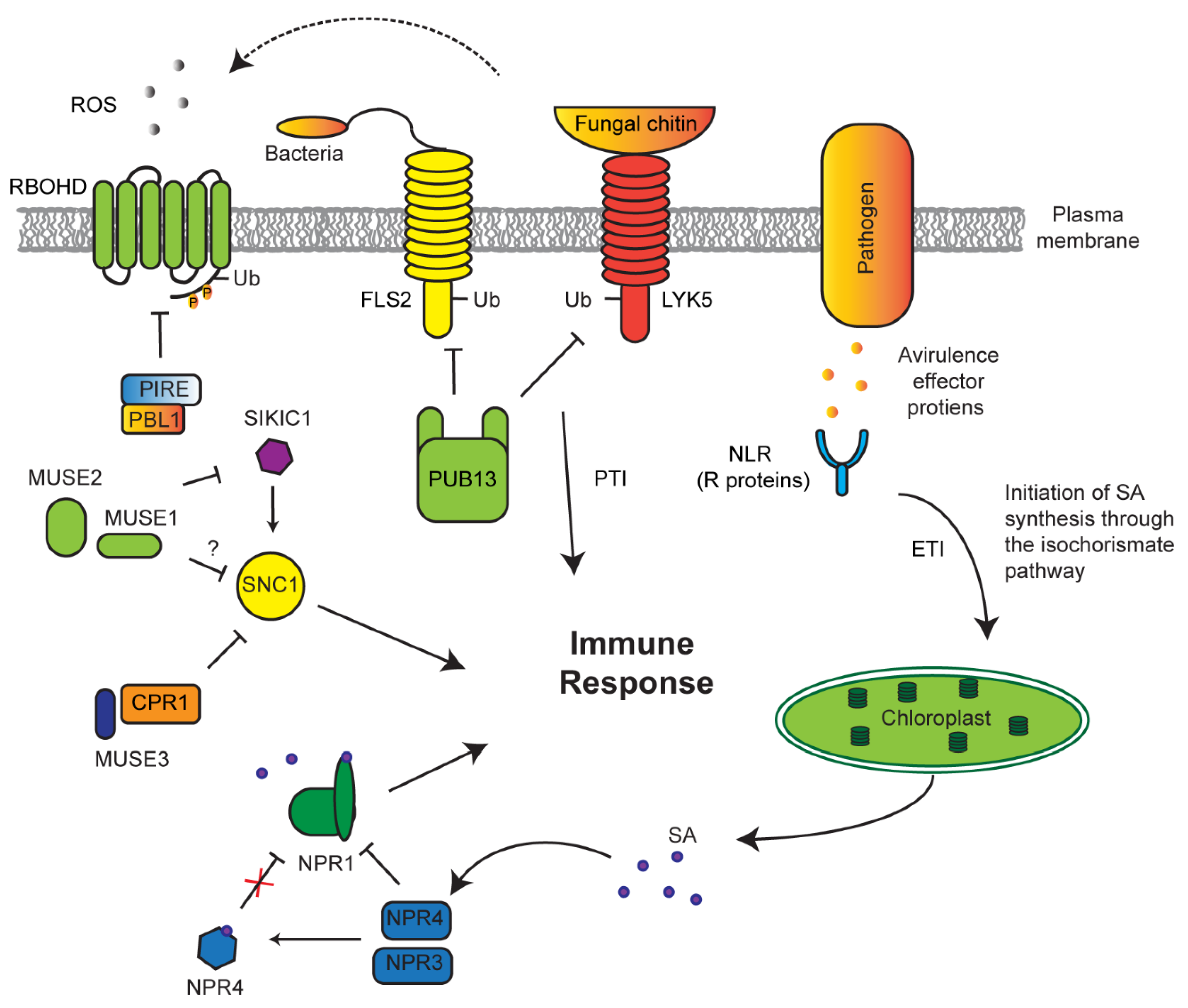
7. Role of the UPS in Innate Immune Response
8. Genome Evolution of the Plant UPS and Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Galmés, J.; Pou, A.; Alsina, M.M.; Tomás, M.; Medrano, H.; Flexas, J. Aquaporin expression in response to different water stress intensities and recovery in Richter-110 (Vitis sp.): Relationship with ecophysiological status. Planta 2007, 226, 671–681. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Mahé, A.; Brangeon, J.; Prioul, J.-L. A maize vacuolar invertase, IVR2, is induced by water stress. Organ/Tissue specificity and diurnal modulation of expression. Plant. Physiol. 2000, 124, 71–84. [Google Scholar] [CrossRef]
- Hua, Z.; Vierstra, R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant. Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26s proteasome proteolytic pathway. Annu. Rev. Plant. Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin–26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Voges, D.; Zwickl, P.; Baumeister, W. The 26s proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999, 68, 1015–1068. [Google Scholar] [CrossRef]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The ubiquitin–proteasome system of saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef]
- Callis, J.; Pollmann, L.; Shanklin, J.; Wettern, M.; Vierstra, R.D. Sequence of a cDNA from Chlamydomonas reinhardii encoding a ubiquitin 52 amino acid extension protein. Nucleic Acids Res. 1989, 17, 8377. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Dynamic regulation of the 26s proteasome: From synthesis to degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Hellmann, H.; Estelle, M. Plant development: Regulation by protein degradation. Science 2002, 297, 793–797. [Google Scholar] [CrossRef]
- Hicke, L.; Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003, 19, 141–172. [Google Scholar] [CrossRef]
- Schwihla, M.; Korbei, B. The beginning of the end: Initial steps in the degradation of plasma membrane proteins. Front. Plant. Sci. 2020, 11. [Google Scholar] [CrossRef]
- Li, W.; Schmidt, W. Non-proteolytic protein ubiquitination is crucial for iron deficiency signaling. Plant. Signal. Behav. 2010, 5, 561–563. [Google Scholar] [CrossRef][Green Version]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. Conventional and unconventional ubiquitination in plant immunity. Mol. Plant. Pathol. 2017, 18, 1313–1330. [Google Scholar] [CrossRef]
- Bard, J.A.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Farmer, L.M.; Book, A.J.; Lee, K.-H.; Lin, Y.-L.; Fu, H.; Vierstra, R.D. The RAD23 family provides an essential connection between the 26s proteasome and ubiquitylated proteins in arabidopsis. Plant. Cell 2010, 22, 124–142. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in arabidopsis. Plant. Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef]
- Hua, Z.; Yu, P. Diversifying evolution of the ubiquitin-26s proteasome system in brassicaceae and poaceae. Int. J. Mol. Sci. 2019, 20, 3226. [Google Scholar] [CrossRef]
- Hua, Z. Diverse evolution in 111 plant genomes reveals purifying and dosage balancing selection models for f-box genes. Int. J. Mol. Sci. 2021, 22, 871. [Google Scholar] [CrossRef]
- Thomas, J.H. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006, 16, 1017–1030. [Google Scholar] [CrossRef]
- Skaar, J.R.; Pagan, J.; Pagano, M. SnapShot: F box proteins I. Cell 2009, 137, 1160–1160.e1. [Google Scholar] [CrossRef]
- Skaar, J.R.; D’Angiolella, V.; Pagan, J.; Pagano, M. SnapShot: F Box Proteins II. Cell 2009, 137, 1358.e1–1358.e2. [Google Scholar] [CrossRef]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant. Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef]
- Freemont, P.S. RING for destruction? Curr. Biol. 2000, 10, R84–R87. [Google Scholar] [CrossRef]
- Mazzucotelli, E.; Belloni, S.; Marone, D.; De Leonardis, A.; Guerra, D.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef]
- Downes, B.P.; Stupar, R.M.; Gingerich, D.J.; Vierstra, R.D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant. J. 2003, 35, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Ohi, M.D.; Kooi, C.W.V.; Rosenberg, J.A.; Chazin, W.J.; Gould, K.L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Mol. Biol. 2003, 10, 250–255. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant e3 ligases: Flexible enzymes in a sessile world. Mol. Plant. 2013, 6, 1388–1404. [Google Scholar] [CrossRef]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Guo, L.; Nezames, C.D.; Sheng, L.; Deng, X.; Wei, N. Cullin-ring ubiquitin ligase family in plant abiotic stress pathwaysf. J. Integr. Plant. Biol. 2013, 55, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; De Veylder, L. The anaphase-promoting complex/Cyclosome in control of plant development. Mol. Plant. 2012, 5, 1182–1194. [Google Scholar] [CrossRef] [PubMed]
- Eloy, N.B.; Lima, M.D.F.; Ferreira, P.C.G.; Inzé, D. The role of the anaphase-promoting complex/Cyclosome in plant growth. Crit. Rev. Plant. Sci. 2015, 34, 487–505. [Google Scholar] [CrossRef]
- Adams, E.H.G.; Spoel, S. The ubiquitin–proteasome system as a transcriptional regulator of plant immunity. J. Exp. Bot. 2018, 69, 4529–4537. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors npr1 and npr3/Npr4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef]
- van Wersch, S.; Tian, L.; Hoy, R.; Li, X. Plant NLRs: The whistleblowers of plant immunity. Plant. Commun. 2020, 1, 1–18. [Google Scholar] [CrossRef]
- Yang, L.; Wu, L.; Chang, W.; Li, Z.; Miao, M.; Li, Y.; Yang, J.; Liu, Z.; Tan, J. Overexpression of the maize E3 ubiquitin ligase gene ZmAIRP4 enhances drought stress tolerance in Arabidopsis. Plant. Physiol. Biochem. 2018, 123, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 are homologous u-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant. Cell 2008, 20, 1899–1914. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, K.; Cheng, Q.; Kong, D.; Zhang, X.; Wang, Z.; Wang, Q.; Xie, Q.; Yan, J.; Chu, J.; et al. Cysteine protease RD21A regulated by E3 ligase SINAT4 is required for drought-induced resistance to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 2020, 71, 5562–5576. [Google Scholar] [CrossRef]
- Min, H.J.; Jung, Y.J.; Kang, B.G.; Kim, A.W.T. CAPUB1, a hot pepper U-box E3 ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice (Oryza sativa L.). Mol. Cells 2016, 39, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.Y.; Cho, S.K.; Kim, W.T. The arabidopsis C3H2C3-type RING e3 ubiquitin ligase ATAIRP1 is a positive regulator of an ABSCISIC acid-dependent response to drought stress. Plant. Physiol. 2010, 154, 1983–1997. [Google Scholar] [CrossRef]
- Yang, R.; Wang, T.; Shi, W.; Li, S.; Liu, Z.; Wang, J.; Yang, Y. E3 ubiquitin ligase ATL61 acts as a positive regulator in abscisic acid mediated drought response in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 528, 292–298. [Google Scholar] [CrossRef]
- Ruan, W.; Guo, M.; Wang, X.; Guo, Z.; Xu, Z.; Xu, L.; Zhao, H.; Sun, H.; Yan, C.; Yi, K. Two RING-Finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant. 2019, 12, 1060–1074. [Google Scholar] [CrossRef]
- Xu, G.; Chen, W.; Song, L.; Chen, Q.; Zhang, H.; Liao, H.; Zhao, G.; Lin, F.; Zhou, H.; Yu, F. Feronia phosphorylates E3 ubiquitin ligase ATL6 to modulate the stability of 14-3-3 proteins in response to the carbon/Nitrogen ratio. J. Exp. Bot. 2019, 70, 6375–6388. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, S.; Sun, H.; Pei, L.; Liu, Y.; Peng, L.; Gao, X.; Liu, Y.; Wang, J. AtPPRT1, an E3 ubiquitin ligase, enhances the thermotolerance in arabidopsis. Plants 2020, 9, 1074. [Google Scholar] [CrossRef]
- Morimoto, K.; Ohama, N.; Fukao, Y.; Fujiwara, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Chen, P.; Zhang, H.; Huang, X.; Zang, Y.; Li, J.; Li, J.; Wong, J. Regulation of ubiquitin-like with plant homeodomain and RING finger domain 1 (UHRF1) protein stability by heat shock protein 90 chaperone machinery. J. Biol. Chem. 2016, 291, 20125–20135. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Doroodian, P.; Vu, W. Contrasting duplication patterns reflect functional diversities of ubiquitin and ubiquitin-like protein modifiers in plants. Plant. J. 2018, 95, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant. Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant. Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant. Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Signorelli, S.; Tarkowski, Ł.P.; Van Den Ende, W.; Bassham, D.C. Linking autophagy to abiotic and biotic stress responses. Trends Plant. Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, W.; Wen, X.; Bulinski, P.J.; Chomchai, D.A.; Arines, F.M.; Liu, Y.-Y.; Sprenger, S.; Teis, D.; Klionsky, D.J.; et al. TORC1 regulates vacuole membrane composition through ubiquitin- and ESCRT-dependent microautophagy. J. Cell Biol. 2020, 219, 201902127. [Google Scholar] [CrossRef]
- Arines, F.M.; Hamlin, A.J.; Yang, X.; Liu, Y.-Y.J.; Li, M. A selective transmembrane recognition mechanism by a membrane-anchored ubiquitin ligase adaptor. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef]
- Tian, M.; Xie, Q. Non-26s proteasome proteolytic role of ubiquitin in plant endocytosis and endosomal traffickingf. J. Integr. Plant. Biol. 2013, 55, 54–63. [Google Scholar] [CrossRef]
- Ling, Q.; Huang, W.; Baldwin, A.; Jarvis, R.P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 2012, 338, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-binding uim proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, W.-Y.; Park, H.C.; Lee, S.Y.; Bohnert, H.J.; Yun, D.-J. SUMO and SUMOylation in plants. Mol. Cells 2011, 32, 305–316. [Google Scholar] [CrossRef]
- Morrell, R.; Sadanandom, A. Dealing with stress: A Review of plant SUMO proteases. Front. Plant. Sci. 2019, 10, 1122. [Google Scholar] [CrossRef]
- Augustine, R.C.; Vierstra, R.D. SUMOylation: Re-wiring the plant nucleus during stress and development. Curr. Opin. Plant. Biol. 2018, 45, 143–154. [Google Scholar] [CrossRef]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural basis for e2-mediated sumo conjugation revealed by a complex between ubiquitin-conjugating enzyme UBC9 and RANGAP1. Cell 2002, 108, 345–356. [Google Scholar] [CrossRef]
- Johnson, E.S.; Blobel, G. Cell cycle-regulated attachment of the ubiquitin-related protein sumo to the yeast septins. J. Cell Biol. 1999, 147, 981–994. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef]
- Miller, M.J.; Scalf, M.; Rytz, T.C.; Hubler, S.L.; Smith, L.M.; Vierstra, R.D. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced sumoylation in arabidopsis. Mol. Cell Proteom. 2013, 12, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Sadanandom, A. An insight into the factors influencing specificity of the sumo system in plants. Plants 2020, 9, 1788. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.J.; Vierstra, R.D. Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant. Signal. Behav. 2011, 6, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Denuc, A.; Marfany, G. SUMO and ubiquitin paths converge. Biochem. Soc. Trans. 2010, 38, 34–39. [Google Scholar] [CrossRef]
- Pan, W.; Lin, B.; Yang, X.; Liu, L.; Xia, R.; Li, J.; Wu, Y.; Xie, Q. The UBC27-AIRP3 ubiquitination complex modulates ABA signaling by promoting the degradation of ABI1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 27694–27702. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Q.; Liu, Z.; Yang, H.; Wang, J.; Liang, Y.; Yang, Y. Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J. Integr. Plant. Biol. 2016, 58, 67–80. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, L.; Gao, X.; Liu, Y.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. AtPPRT3, a novel E3 ubiquitin ligase, plays a positive role in ABA signaling. Plant. Cell Rep. 2020, 39, 1467–1478. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor Lysin motif receptor kinase5 (LYK5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef]
- Seo, D.H.; Ryu, M.Y.; Jammes, F.; Hwang, J.H.; Turek, M.; Kang, B.G.; Kwak, J.M.; Kim, W.T. Roles of four arabidopsis U-BOX E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant. Physiol. 2012, 160, 556–568. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Wu, Y.-R.; Huang, X.-H.; Sun, J.; Xie, Q. ATPUB 19, a U-BOX E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in arabidopsis thaliana. Mol. Plant. 2011, 4, 938–946. [Google Scholar] [CrossRef]
- Dong, O.X.; Ao, K.; Xu, F.; Johnson, K.C.M.; Wu, Y.; Li, L.; Xia, S.; Liu, Y.; Huang, Y.; Rodriguez, E.; et al. Individual components of paired typical NLR immune receptors are regulated by distinct E3 ligases. Nat. Plants 2018, 4, 699–710. [Google Scholar] [CrossRef]
- Zhang, N.; Yin, Y.; Hu, Z.; Ni, Z.; Sun, Q.; Yao, Y.; Liu, X.; Tong, S.; Xing, J.; Zhang, Y.; et al. The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant. Physiol. 2017, 175, 1878–1892. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Tran, L.-S.P.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant. Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, G.; Yan, J.; He, C.; Zhang, H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant. J. 2006, 46, 649–657. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, H.-L.; Li, H.-G.; Su, Y.; Li, S.; Yang, Y.; Feng, C.-H.; Yin, W.; Xia, X. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant. Biotechnol. J. 2018, 16, 1514–1528. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nat. Cell Biol. 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Gou, M.; Shi, Z.; Zhu, Y.; Bao, Z.; Wang, G.; Hua, J. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant. J. 2011, 69, 411–420. [Google Scholar] [CrossRef]
- Zhu, Z.; Xu, F.; Zhang, Y.; Cheng, Y.T.; Wiermer, M.; Li, X. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. USA 2010, 107, 13960–13965. [Google Scholar] [CrossRef]
- Marino, D.; Froidure, S.; Canonne, J.; Ben Khaled, S.; Khafif, M.; Pouzet, C.; Jauneau, A.; Roby, D.; Rivas, S. Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 2013, 4, 1476. [Google Scholar] [CrossRef]
- Huang, Y.; Minaker, S.; Roth, C.; Huang, S.; Hieter, P.; Lipka, V.; Wiermer, M.; Li, X. An E4 ligase facilitates polyubiquitination of plant immune receptor resistance proteins in arabidopsis. Plant. Cell 2014, 26, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Withers, J.; Li, H.; Zwack, P.J.; Rusnac, D.-V.; Shi, H.; Liu, L.; Yan, S.; Hinds, T.R.; Guttman, M.; et al. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nat. Cell Biol. 2020, 586, 311–316. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nat. Cell Biol. 2012, 486, 228–232. [Google Scholar] [CrossRef]
- Lee, D.; Lal, N.K.; Lin, Z.-J.D.; Ma, S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.-J.; Ma, L.; Liao, C. The ubiquitin E3 ligase RHA2b promotes degradation of MYB30 in abscisic acid signaling. Plant. Physiol. 2018, 178, 428–440. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant. Sci. 2020, 11. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Sharma, S.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant. Sci. 2017, 8, 161. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant. Sci. 2016, 7, 571. [Google Scholar] [CrossRef]
- Shu, K.; Luo, X.; Meng, Y.; Yang, W. Toward a molecular understanding of abscisic acid actions in floral transition. Plant. Cell Physiol. 2018, 59, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, W.; Wang, X.-X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant. Biotechnol. J. 2016, 15, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC acid-insensitive 2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant. Cell 1997, 9, 759–771. [Google Scholar] [PubMed]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant. Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef]
- Wu, Y.; Sanchez, J.P.; Lopez-Molina, L.; Himmelbach, A.; Grill, E.; Chua, N.H. The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant. J. 2003, 34, 307–315. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N.; Giraudat, J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant. Cell 1999, 11, 1897–1910. [Google Scholar]
- Ali, A.; Pardo, J.M.; Yun, D.-J. Desensitization of aba-signaling: The swing from activation to degradation. Front. Plant. Sci. 2020, 11, 379. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.; Rodriguez, P.L.; An, C. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant. Cell 2016, 28, 2178–2196. [Google Scholar] [CrossRef]
- Belda-Palazon, B.; Julian, J.; Coego, A.; Wu, Q.; Zhang, X.; Batistic, O.; AlQuraishi, S.A.; Kudla, J.; An, C.; Rodriguez, P.L. ABA inhibits myristoylation and induces shuttling of the RGLG 1 E3 ligase to promote nuclear degradation of PP 2 CA. Plant. J. 2019, 98, 813–825. [Google Scholar] [CrossRef]
- Meinnel, T.; Dian, C.; Giglione, C. Myristoylation, an ancient protein modification mirroring eukaryogenesis and evolution. Trends Biochem. Sci. 2020, 45, 619–632. [Google Scholar] [CrossRef]
- Jimenez-Quesada, M.J.; Meinnel, T.; Giglione, C. Expanded impact of protein N-myristoylation in plants. Plant. Signal. Behav. 2008, 3, 501–502. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nat. Cell Biol. 2005, 435, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nat. Cell Biol. 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nat. Cell Biol. 2007, 446, 640–645. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.G.D.C.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garciacasado, G.; Lopezvidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nat. Cell Biol. 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.D.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nat. Cell Biol. 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef]
- Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Seki, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant. Mol. Biol. 2000, 42, 657–665. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant. Cell 1998, 10, 1391–1406. [Google Scholar]
- Morimoto, K.; Mizoi, J.; Qin, F.; Kim, J.-S.; Sato, H.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Stabilization of arabidopsis DREB2A is required but not sufficient for the induction of target genes under conditions of stress. PLoS ONE 2013, 8, e80457. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant. Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Y.; Lai, J.; Yang, C.; Shi, Y.; Han, D.; Wu, Y.; Ye, W.; Yang, H.; Li, G.; et al. SUMOylation stabilizes the transcription factor DREB2A to improve plant thermotolerance. Plant. Physiol. 2020, 183, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Qu, G.-P.; Kong, X.; Yan, Y.; Li, J.; Jin, J.B. Arabidopsis small ubiquitin-related modifier protease ASP1 positively regulates abscisic acid signaling during early seedling development. J. Integr. Plant. Biol. 2018, 60, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lai, J.; Du, J.; Xie, Q.; Wu, K.; Yang, C.; Wang, F.; Yang, S.; He, Z.; Jiang, J.; et al. A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant. Physiol. 2017, 173, 1574–1582. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Kangasjärvi, J. ROS signaling loops-production, perception, regulation. Curr. Opin. Plant. Biol. 2013, 16, 575–582. [Google Scholar] [CrossRef]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant. Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Vogel, M.O.; Moore, M.; König, K.; Pecher, P.; Alsharafa, K.; Lee, J.; Dietz, K.-J. Fast retrograde signaling in response to high light involves metabolite export, mitogen-activated protein kinase6, and AP2/ERF transcription factors in arabidopsis[C][w]. Plant. Cell 2014, 26, 1151–1165. [Google Scholar] [CrossRef]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef]
- Sierla, M.; Waszczak, C.; Vahisalu, T.; Kangasjärvi, J. Reactive oxygen species in the regulation of stomatal movements. Plant. Physiol. 2016, 171, 1569–1580. [Google Scholar] [CrossRef]
- Kim, T.-H.; Böhmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and CA2+signaling. Annu. Rev. Plant. Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, D.B.; Barros, J.A.; Fernie, A.R.; Araújo, W.L. Eating away at ROS to regulate stomatal opening. Trends Plant. Sci. 2020, 25, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Murata, Y.; Benning, G.; Thomine, S.; Klusener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Rahikainen, M.; Pascual, J.; Alegre, S.; Durian, G.; Kangasjärvi, S. PP2A phosphatase as a regulator of ros signaling in plants. Antioxidants 2016, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. Ubiquitin goes green. Trends Cell Biol. 2016, 26, 3–5. [Google Scholar] [CrossRef]
- Woodson, J.D.; Joens, M.S.; Sinson, A.B.; Gilkerson, J.; Salomé, P.A.; Weigel, D.; Fitzpatrick, J.A.; Chory, J. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 2015, 350, 450–454. [Google Scholar] [CrossRef]
- Ling, Q.; Jarvis, P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 2015, 25, 2527–2534. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nat. Cell Biol. 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Menezes, H.; Jared, C. Immunity in plants and animals: Common ends through different means using similar tools. Comp. Biochem. Physiol. Part. C Toxicol. Pharmacol. 2002, 132, 1–7. [Google Scholar] [CrossRef]
- Han, G.-Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nat. Cell Biol. 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Duplan, V.; Rivas, S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant. Sci. 2014, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Peeters, N.; Rivas, S. Ubiquitination during plant immune signaling. Plant. Physiol. 2012, 160, 15–27. [Google Scholar] [CrossRef]
- Devoto, A.; Muskett, P.R.; Shirasu, K. Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant. Biol. 2003, 6, 307–311. [Google Scholar] [CrossRef]
- Craig, A.; Ewan, R.; Mesmar, J.; Gudipati, V.; Sadanandom, A. E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot. 2009, 60, 1123–1132. [Google Scholar] [CrossRef]
- Dong, X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant. Biol. 1998, 1, 316–323. [Google Scholar] [CrossRef]
- Miricescu, A.; Goslin, K.; Graciet, E. Ubiquitylation in plants: Signaling hub for the integration of environmental signals. J. Exp. Bot. 2018, 69, 4511–4527. [Google Scholar] [CrossRef]
- Kelley, D.R. E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol. Cell Proteom. 2018, 17, 1047–1054. [Google Scholar] [CrossRef]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef]
- Dodds, P.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant. Pathol. 2011, 12, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zeng, L. The tomato U-BOX type E3 ligase PUB13 acts with group III ubiquitin E2enzymes to modulate FLS2-mediated immune signaling. Front. Plant. Sci. 2018, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Li, X.; Kapos, P.; Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 2015, 32, 114–121. [Google Scholar] [CrossRef]
- Cheng, Y.T.; Li, Y.; Huang, S.; Huang, Y.; Dong, X.; Zhang, Y.; Li, X. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 14694–14699. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant. Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef]
- Lin, Z.-J.D.; Liebrand, T.W.H.; Yadeta, K.A.; Coaker, G. PBL13 is a serine/Threonine protein kinase that negatively regulates arabidopsis immune responses. Plant. Physiol. 2015, 169, 2950–2962. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant. J. 2007, 51, 941–954. [Google Scholar] [CrossRef]
- Raffaele, S.; Vailleau, F.; Léger, A.; Joubès, J.; Miersch, O.; Huard, C.; Blée, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in arabidopsis. Plant. Cell 2008, 20, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.-L.; Triantaphylidès, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.Y.; Xue, H.W. PI4Kgamma2 interacts with E3 ligase MIEL1 to regulate auxin metabolism and root development. Plant. Physiol. 2020, 184, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB. Nat. Commun. 2016, 7, 12525. [Google Scholar]
- Li, Y.; Yapa, M.M.; Hua, Z. A machine-learning approach to prioritizing active F-box members in Arabidopsis thaliana. Front. Plant. Sci. 2021. in review. [Google Scholar]
- Hua, Z.; Pool, J.E.; Schmitz, R.J.; Schultz, M.D.; Shiu, S.-H.; Ecker, J.R.; Vierstra, R.D. Epigenomic programming contributes to the genomic drift evolution of the F-Box protein superfamily in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 16927–16932. [Google Scholar] [CrossRef]
- Hua, Z.; Zou, C.; Shiu, S.-H.; Vierstra, R.D. Phylogenetic comparison of F-box (FBX) Gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE 2011, 6, e16219. [Google Scholar] [CrossRef]



| Species | BTB | F-box * | HECT | RING ** | Sum |
|---|---|---|---|---|---|
| Arabidopsis halleri | 63 | 850 | 9 | 480 | 1402 |
| Arabidopsis lyrate | 77 | 989 | 10 | 542 | 1618 |
| Arabidopsis thaliana | 64 | 697 | 7 | 516 | 1284 |
| Amborella tricopoda | 56 | 230 | 7 | 314 | 607 |
| Brachypodium distachyon | 178 | 813 | 10 | 554 | 1555 |
| Brassica rapa | 97 | 975 | 10 | 802 | 1884 |
| Boechera stricta | 75 | 505 | 9 | 473 | 1062 |
| Capsella rubella | 69 | 970 | 8 | 527 | 1574 |
| Leersia perrieri | 110 | 542 | 9 | 460 | 1121 |
| Oryza brachyantha | 78 | 264 | 12 | 391 | 745 |
| Oryza punctata | 111 | 535 | 8 | 485 | 1139 |
| Oryza sativa | 156 | 732 | 8 | 532 | 1428 |
| Sorghum bicolor | 158 | 678 | 9 | 563 | 1408 |
| Zea mays | 95 | 331 | 20 | 699 | 1145 |
| Protein | Accession | Involved Stress Pathway (Substrates) | Protein Family | Reference |
|---|---|---|---|---|
| ABA Signaling | ||||
| AIRP1 | AT4G23450.2 | Positive regulator in ABA-mediated drought tolerance | RING E3 ligase | [46] |
| AIRP3 | AT3G09770.1 | E3 ligase for ABI1 degradation (ABI1) | RING E3 ligase | [75] |
| AtAIRP4 | AT2G36270.1 | Positive regulator in ABA-mediated drought tolerance | RING E3 ligase | [76] |
| AtL61 | AT3G14320.1 | Positive regulator in ABA-mediated drought tolerance | RING-H2 E3 ligase | [47] |
| AtPPRt3 | AT1G18470.1 | Positive regulator in ABA-mediated drought tolerance | RING E3 ligase | [77] |
| PUB12 | AT2G28830.1 | Drought and immune response (ABI1) | U-box E3 ligase | [78] |
| PUB13 | AT3G46510.1 | Drought and immune response (ABI1) | U-box E3 ligase | [79] |
| PUB18 | AT1G10560.1 | ABA signaling | U Box E3 ligase | [80] |
| PUB19 | AT1G60190.1 | Negative regulator of ABA-mediated drought tolerance | U-box E3 ligase | [81] |
| RGLG1 | AT3G01650.1 | Positive regulator of ABA-mediated drought tolerance (PP2CA) | RING E3 ligase | [82] |
| UBC27 | AT5G50870.2 | E2 that conjugates with ABI1 | E2 | [75] |
| DREB2A-Mediated Stress Tolerance | ||||
| BPM | AT1G21780.1 | Thermotolerance (DREB2A) | CRL-BTB E3 ligase | [51] |
| SAP5 | AT3G12630.1 | Thermotolerance (DRIP1/2) | A20/AN1 domain containing E3 ligase | [83] |
| DRIP1/2 | AT1G06770.1 | Thermotolerance (DREB2A) | RING E3 ligase | [84] |
| ROS Homeostasis | ||||
| AtCHIP | AT3G07370.1 | Regulator of temperature response (PP2A) | U Box E3 ligase | [85] |
| PeCHYR1 | AT5G22920.1 | Positive regulator in ABA-mediated drought tolerance | Ring E3 ligase | [86] |
| SP1 | AT1G63900.2 | Chloroplast biogenesis and ROS homeostasis (TOC complex) | RING E3 ligase | [61] |
| Innate Immune Response | ||||
| COI1 | AT2G39940.1 | Jasmonic acid receptor (JAZ proteins) | SCF CRL E3 ligase | [87] |
| CPR1 | AT4G12560.1 | Negative regulator of immunity (SNC1) | SCF CRL E3 ligase | [88,89] |
| MIEL1 | AT5G18650.1 | Immune response (MYB30) | RING E3 ligase | [90] |
| MUSE1 | AT3G58030.1 | Regulates SIKIC1 (SIKIC1) | RING E3 ligase | [82] |
| MUSE2 | AT2G42030.1 | Regulates SIKIC1 (SIKIC1) | RING E3 ligase | [82] |
| MUSE3 | AT5G15400.1 | Mediates SNC1 ubiquitylation | E4 ligase | [91] |
| NPR1 | AT1G64280.1 | SA receptor | CRL-BTB E3 ligase | [92,93,94] |
| NPR3/4 | AT5G45110.1 | SA co-receptor (NPR1) | CRL-BTB E3 ligase | [95] |
| PIRE | RBOHD degradation (RBOHD) | RING E3 ligase | [96] | |
| PUB12 | AT2G28830.1 | Drought and immune response (FLS2) | U-box E3 ligase | [78] |
| PUB13 | AT3G46510.1 | Drought and immune response (LYK5) | U-box E3 ligase | [79] |
| RHA2B | AT2G01150.1 | Immune response (MYB30) | RING E3 ligase | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doroodian, P.; Hua, Z. The Ubiquitin Switch in Plant Stress Response. Plants 2021, 10, 246. https://doi.org/10.3390/plants10020246
Doroodian P, Hua Z. The Ubiquitin Switch in Plant Stress Response. Plants. 2021; 10(2):246. https://doi.org/10.3390/plants10020246
Chicago/Turabian StyleDoroodian, Paymon, and Zhihua Hua. 2021. "The Ubiquitin Switch in Plant Stress Response" Plants 10, no. 2: 246. https://doi.org/10.3390/plants10020246
APA StyleDoroodian, P., & Hua, Z. (2021). The Ubiquitin Switch in Plant Stress Response. Plants, 10(2), 246. https://doi.org/10.3390/plants10020246






