Changes in the Plant β-Sitosterol/Stigmasterol Ratio Caused by the Plant Parasitic Nematode Meloidogyne incognita
Abstract
:1. Introduction
2. Results and Discussion
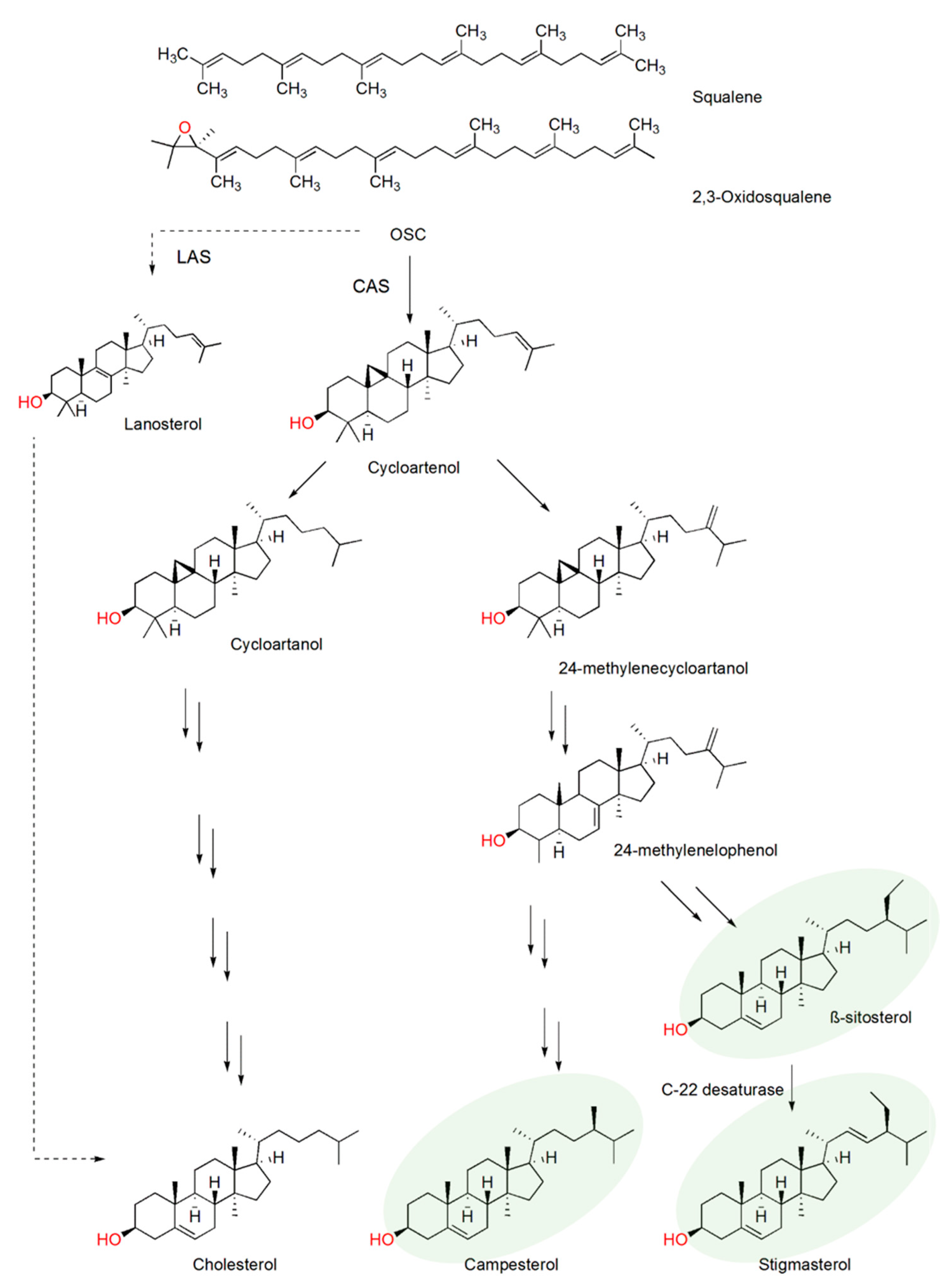
2.1. Plant Sterol Composition
2.2. Plant Sterol Composition after Meloidogyne Incognita Infection
2.3. β-Sitosterol/Stigmasterol Conversion in Tomato after Meloidogyne Incognita Infection
2.4. CYP710A
3. Materials and Methods
3.1. Nematode Inoculation and Plant Material
3.2. Sterol Extraction and GC-MS Analysis
3.3. CYP710A11 Temporal Gene Expression Analysis
3.4. Phylogenetic Analysis of Cytochrome P450 Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- London, E. Insights into lipid raft structure and formation from experiments in model membranes. Curr. Opin. Struct. Biol. 2002, 12, 480–486. [Google Scholar] [CrossRef]
- Arnqvist, L.; Persson, M.; Jonsson, L.; Dutta, P.C.; Sitbon, F. Overexpression of CYP710A1 and CYP710A4 in transgenic Arabidopsis plants increases the level of stigmasterol at the expense of sitosterol. Planta 2007, 227, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Sewelam, N.; Jaspert, N.; Van Der Kelen, K.; Tognetti, V.B.; Scmitz, J.; Frerigmann, H.; Stahl, E.; Zeier, J.; Van Breusege, F.; Maurino, V.G. Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 2014, 7, 1191–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valitova, J.N.; Sulkarnayeva, A.G.; Minibayeva, F.V. Plant sterols: Diversity, biosynthesis, and physiological functions. Biochemistry 2016, 81, 1050–1068. [Google Scholar] [CrossRef] [PubMed]
- Aboobucker, S.I.; Suza, W.P. Why do plants convert sitosterol to stigmasterol? Front. Plant Sci. 2019, 10, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmond, E.; Gribaldo, S. Phylogenomics of sterol synthesis: Insights into the origin, evolution and diversity of a key eukaryotic feature. Genome Biol. Evol. 2009, 1, 364–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebedev, R.; Trabelcy, B.; Goncalves, I.L.; Gerchman, Y.; Sapir, A. Metabolic reconfiguration in C. elegans suggests a pathway for widespread sterol auxotrophy in the animal kingdom. Curr. Biol. 2020, 30, 3031–3038. [Google Scholar] [CrossRef]
- Chitwood, D.J.; Lusby, W.R. Metabolism of plant sterols by nematodes. Lipids 1991, 26, 619–627. [Google Scholar] [CrossRef]
- Chitwood, D.J. Biochemistry and functions of nematode steroids. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 273–284. [Google Scholar] [CrossRef]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveal complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Kadota, Y.; Shirasu, K. Plant immune responses to parasitic nematodes. Front. Plant Sci. 2019, 10, 1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, M.A. Plant sterols and the membrane environment. Trends Plant Sci. 1998, 3, 170–175. [Google Scholar] [CrossRef]
- Hodzic, A.; Rappolt, M.; Amenitsch, H.; Laggner, P.; Pabst, G. Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys. J. 2008, 94, 3935–3944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griebel, T.; Zeier, J. A role for β-sitosterol to stigmasterol conversion in plant-pathogen interaction. Plant J. 2010, 63, 254–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Senthil-Kumar, M.; Ryu, C.; Kang, L.; Mysore, K.S. Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol. 2012, 158, 1789–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinovieva, S.V.; Vasyukova, N.I.; Ozeretskovskaya, I.L. Involvement of plant sterols in the system tomatoes-nematode Meloidogyne incognita. Helminthologia 1990, 27, 211–216. [Google Scholar]
- Hedin, P.A.; Callahan, F.E.; Dollar, D.A.; Greech, R.G. Total sterols in root-knot nematode Meloidogyne incognita infected cotton Gossypium hirsutum (L.) plant roots. Comp. Biochem. Physiol. 1995, 111, 447–452. [Google Scholar] [CrossRef]
- Sawai, S.; Ohyama, K.; Yasumoto, S.; Seki, H.; Sakuma, T.; Yamamoto, T.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; Aoki, T.; et al. Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 2014, 26, 3763–3774. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, T.; Mizutani, M.; Aoki, N.; Watanabe, B.; Saga, H.; Saito, S.; Oikawa, A.; Suzuki, H.; Sakurai, N.; Shibata, D.; et al. Cytochrome P450 CYP710A encodes the sterol C-22 desaturase in Arabidopsis and tomato. Plant Cell 2006, 18, 1008–1022. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.R. Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 2006, 5, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Piironen, V.; Toivo, J.; Puupponen-Pimiä, R.; Lampi, A.M. Plant sterols in vegetables, fruits and berries. J. Sci. Food Agric. 2003, 83, 330–337. [Google Scholar] [CrossRef]
- Jun-Hua, H.A.N.; Yue-Xin, Y.A.N.G.; Mei-Yuan, F.E.N.G. Contents of phytosterols in vegetables and fruits commonly consumed in China. Biomed. Environ. Sci. 2008, 21, 449–453. [Google Scholar] [CrossRef]
- Jin, D.; Dai, K.; Xie, Z.; Chen, J. Secondary metabolites profiled in cannabis inflorescences, leaves, stem barks, and roots for medicinal purposes. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Bladocha, M.; Benveniste, P. Manipulation by tridemorph, a systemic fungicide, of the sterol composition of maize leaves and roots. Plant Physiol. 1983, 71, 756–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalbi, N.; Martínez-Ballesta, M.C.; Youssef, N.B.; Carvajal, M. Intrinsic stability of Brassicaceae plasma membrane in relation to changes in proteins and lipids as a response to salinity. J. Plant Physiol. 2015, 175, 148–156. [Google Scholar] [CrossRef]
- Surjus, A.; Durand, M. Lipid changes in soybean root membranes in response to salt treatment. J. Exp. Bot. 1996, 47, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Ryan, E.; Galvin, K.; O’Connor, T.P.; Maguire, A.R.; O’Brien, N.M. Phytosterol, squalene, tocopherol content and fatty acid profile of selected seeds, grains, and legumes. Plant Foods Hum. Nutr. 2007, 62, 85–91. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, K.; Li, Y. Highlights to phytosterols accumulation and equilibrium in plants: Biosynthetic pathway and feedback regulation. Plant Physiol. Biochem. 2020, 155, 637–649. [Google Scholar] [CrossRef]
- Ferrer, A.; Altabella, T.; Arró, M.; Boronat, A. Emerging roles for conjugated sterols in plants. Prog. Lipid Res. 2017, 67, 27–37. [Google Scholar] [CrossRef]
- Behmer, S.; Olszewski, N.; Sebastiani, J.; Palka, S.; Sparacino, G.; Grebenok, R.J. Plant phloem sterol content: Forms, putative functions, and implications for phloem-feeding insects. Front. Plant Sci. 2013, 4, 370. [Google Scholar] [CrossRef] [Green Version]
- Petersson, E.V.; Nahar, N.; Dahlin, P.; Broberg, A.; Tröger, R.; Dutta, P.C.; Jonsson, L.; Sitbon, F. Conversion of exogenous cholesterol into glycoalkaloids in potato shoots, using two methods for sterol solubilisation. PLoS ONE 2013, 8, e82955. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A. Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. 2007, 45, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.; Aranda, F.J.; Ortiz, A.; Martínez, V.; Carvajal, M.; Teruel, J.A. Molecular aspects of the interaction between plants sterols and DPPC bilayers: An experimental and theoretical approach. J. Colloid Interface Sci. 2011, 358, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Azadmard-Damirchi, S.; Peighambardoust, S.H.; Hesari, J.; Valizadeh, H.; Faller, R. Molecular dynamics simulations of ternary lipid bilayers containing plant sterol and glucosylceramide. Chem. Phys. Lipids 2017, 203, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.O. Secondary Plant Products; Conn, E.E., Ed.; Academic Press: New York, NY, USA, 1981; Volume 7, pp. 501–525. ISBN 978-0-12-675407-0. [Google Scholar]
- Matthiessen, J.N.; Kirkegaard, J.A. Biofumigation and biodegradation: Opportunity and challenge in soil-borne pest and disease management. Crit. Plant Sci. 2006, 25, 235–265. [Google Scholar] [CrossRef]
- Morris, E.K.; Fletcher, R.; Veresoglou, S.D. Effective methods of biofumigation: A meta-analysis. Plant Soil 2019, 446, 379–392. [Google Scholar] [CrossRef]
- Dahlin, P.; Hallmann, J. New insights on the role of allyl isothiocyanate in controlling the root knot nematode Meloidogyne hapla. Plants 2020, 9, 603. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.; Ploeg, A. Evaluation of 31 potential biofumigant brassicaceous plants as hosts for three Meloiodogyne species. J. Nematol. 2014, 46, 287–295. [Google Scholar]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Rogowska, A.; Szakiel, A. The role of sterols in plant response to abiotic stress. Phytochem. Rev. 2020, 19, 1525–1538. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nat. Rev. 2006, 444, 323–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, P.H.; Zscheile, F.P., Jr.; Brannaman, B.L. Sterol changes in maize leaves infected with Helminthosporium carbonum. Plant Physiol. 1970, 45, 634–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, R.; Kim, W.K.; Rohringer, R. Sterols of healthy and rust-infected primary leaves of wheat and of non-germinated and germinated uredospores of wheat stem rust. Can. J. Bot. 1972, 50, 185–190. [Google Scholar] [CrossRef]
- Kissen, R.; Pope, T.W.; Grant, M.; Pickett, J.A.; Rossiter, J.T.; Powell, G. Modifying the alkylglucosinolate profile in Arabidopsis thaliana alters the tritrophic interaction with the herbivore Brevicoryne brassicae and parasitoid Diaeretiella rapae. J. Chem. Ecol. 2009, 35, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S. Triterpene structural diversification by plant cytochrome P450 enzymes. Front. Plant Sci. 2017, 8, 1886. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Saga, H.; Hashizume, H.; Ohta, D. CYP710A genes encoding sterol C22-desaturase in Physcomitrella patens as molecular evidence for the evolutionary conservation of a sterol biosynthetic pathway in plants. Planta 2009, 229, 1311–1322. [Google Scholar] [CrossRef]
- Ramadan, A.M.; Azeiz, A.A.; Baabad, S.; Hassanein, S.; Gadalla, N.O.; Hassan, S.; Algandaby, M.; Bakr, S.; Khan, T.; Abouseadaa, H.H.; et al. Control of β-sitosterol biosynthesis under light and watering in desert plant Calotropis procera. Steroids 2019, 141, 1–8. [Google Scholar] [CrossRef]
- Yu, J.; Tehrim, S.; Wang, L.; Dossa, K.; Zhang, X.; Ke, T.; Liao, B. Evolutionary history and functional divergence of the cytochrome P450 gene superfamily between Arabidopsis thaliana and Brassica species uncover effects of whole genome and tandem duplications. BMC Genom. 2017, 18, 733. [Google Scholar] [CrossRef] [Green Version]
- PM 7/119 (1). Nematode extraction. EPPO Bull. 2013, 43, 471–495. [Google Scholar] [CrossRef]
- Blight, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Dahlin, P.; Srivastava, V.; Ekengren, S.; McKee, L.S.; Bulone, V. Comparative analysis of sterol acquisition in the oomycetes Saprolegnia parasitica and Phytophthora infestans. PLoS ONE 2017, 12, e0170873. [Google Scholar] [CrossRef] [PubMed]
- Azadmard-Damirchi, S.; Dutta, P.C. Novel solid-phase extraction method to separate 4-desmethyl-, 4-monomethyl- and 4,4′-dimethylsterols in vegetable oils. J. Chromatogr. A 2006, 1108, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putna, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]




| Plant Species | Sample | Cholesterol | Campesterol | Stigmasterol | β-Sitosterol |
|---|---|---|---|---|---|
| B. juncea | Root | 0.1 (0.1) | 4.1 (5.6) | 1.7 (1.6) | 94.1 (92.7) |
| Inf. root | 0.1 (0.2) | 5.6 (7.3) | 1.9 (1.9) | 92.5 (90.7) | |
| p value | 0.79 | 0.07 | 0.72 | 0.12 | |
| C. sativus | Root | ND (0.1) | ND (ND) | 99.7 (99.5) | 0.3 (0.5) |
| Inf. root | ND (0.2) | ND (ND) | 99.0 (99.1) | 1.0 (0.7) | |
| p value | NA | NA | 0.03 * | 0.03 * | |
| G. max | Root | 0.1 (0.3) | 2.6 (2.3) | 62.4 (56.5) | 34.9 (40.9) |
| Inf. root | 0.1 (0.2) | 2.6 (2.5) | 61.7 (59.3) | 35.7 (37.8) | |
| p value | 0.95 | 0.07 | 0.45 | 0.41 | |
| S. lycopersicum cv. Moneymaker | Root | 6.5 (9.1) | 1.9 (2.3) | 86.7 (75.5) | 5.0 (13.1) |
| Inf. root | 7.5 (11.4) | 1.9 (3.0) | 75.0 (65.6) | 15.6 (20.0) | |
| p value | 0.15 | 0.28 | 9.4 × 10−4 *** | 5.1 × 10−5 *** | |
| S. lycopersicum cv. Oskar | Root | 6.1 (7.3) | 1.1 (1.5) | 84.7 (80.3) | 8.0 (10.9) |
| Inf. root | 8.2 (9.8) | 1.5 (1.7) | 78.7 (75.4) | 11.6 (13.2) | |
| p value | 0.1 | 0.02 * | 0.07 | 0.09 | |
| Z. mays | Root | 0.1 (0.2) | 5.3 (5.3) | 82.7 (81.2) | 11.9 (13.3) |
| Inf. root | 0.2 (0.3) | 5.8 (5.5) | 80.7 (80.7) | 13.3 (13.5) | |
| p value | 0.05 * | 0.09 | 0.1 | 0.003 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabianca, A.; Müller, L.; Pawlowski, K.; Dahlin, P. Changes in the Plant β-Sitosterol/Stigmasterol Ratio Caused by the Plant Parasitic Nematode Meloidogyne incognita. Plants 2021, 10, 292. https://doi.org/10.3390/plants10020292
Cabianca A, Müller L, Pawlowski K, Dahlin P. Changes in the Plant β-Sitosterol/Stigmasterol Ratio Caused by the Plant Parasitic Nematode Meloidogyne incognita. Plants. 2021; 10(2):292. https://doi.org/10.3390/plants10020292
Chicago/Turabian StyleCabianca, Alessandro, Laurin Müller, Katharina Pawlowski, and Paul Dahlin. 2021. "Changes in the Plant β-Sitosterol/Stigmasterol Ratio Caused by the Plant Parasitic Nematode Meloidogyne incognita" Plants 10, no. 2: 292. https://doi.org/10.3390/plants10020292
APA StyleCabianca, A., Müller, L., Pawlowski, K., & Dahlin, P. (2021). Changes in the Plant β-Sitosterol/Stigmasterol Ratio Caused by the Plant Parasitic Nematode Meloidogyne incognita. Plants, 10(2), 292. https://doi.org/10.3390/plants10020292








