Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions
Abstract
1. Introduction
2. Results
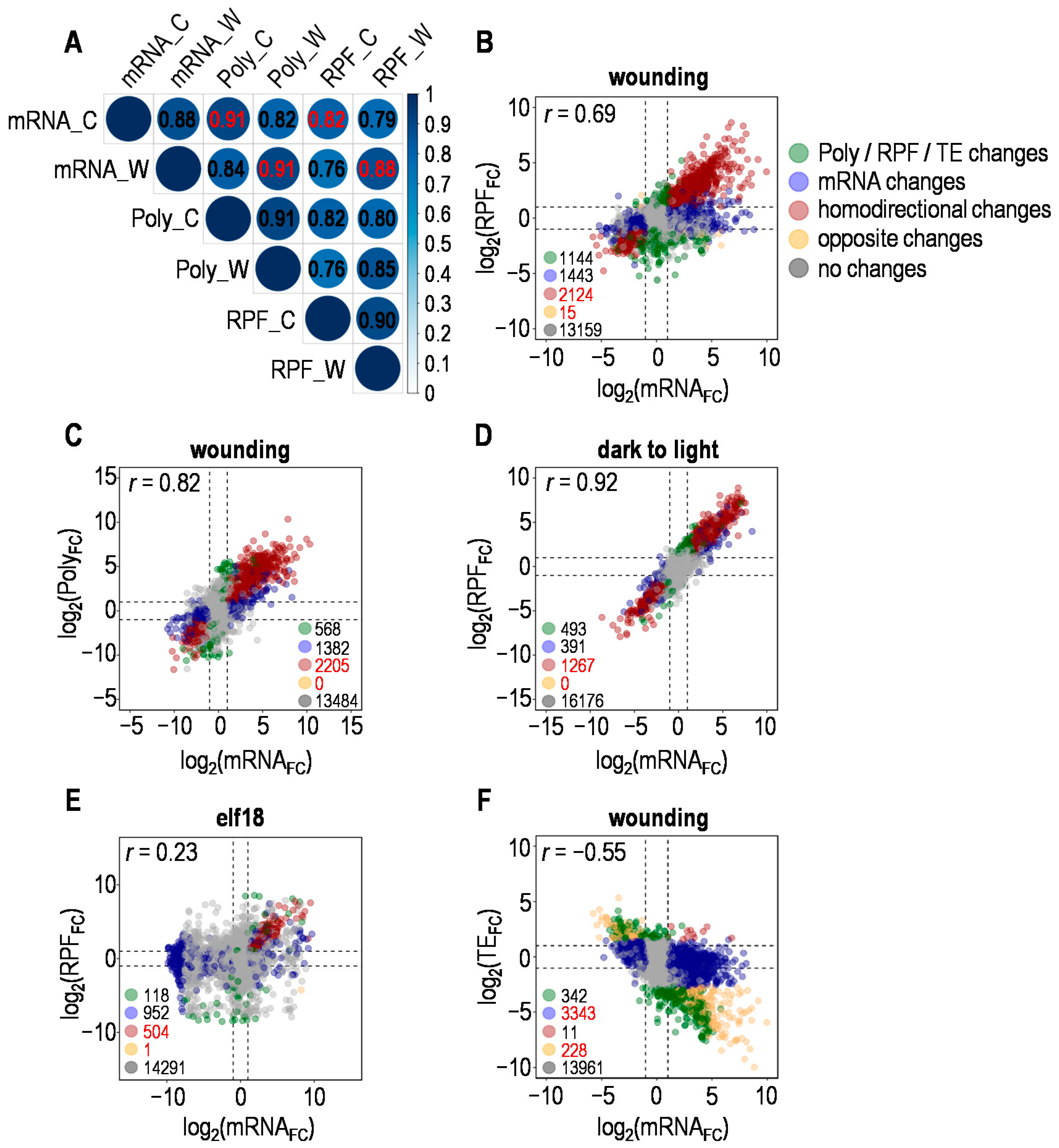
2.1. Stress-Dependent Correlation between Changes in Transcriptome and Translatome
2.2. Divergent and Coordinated Changes in Transcriptome and Translatome upon Wounding
2.3. Targeted Translational Control under Different Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Treatment
4.2. Preparation of Total, Polysome-Associated mRNA and Ribosome-Protected Fragments
4.3. Library Construction and Sequencing
4.4. Bioinformatic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Nadal, E.; Ammerer, G.; Posas, F. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011, 12, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Qin, B.; Nikolay, R.; Spahn, C.M.T.; Zhang, G. Translatomics: The global view of translation. Int. J. Mol. Sci. 2019, 20, 212. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Sina, G.; Newman, J.R.S.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Kuster, B. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Arava, Y.; Wang, Y.; Storey, J.D.; Liu, C.L.; Brown, P.O.; Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 3889–3894. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef]
- Dunn, J.G.; Foo, C.K.; Belletier, N.G.; Gavis, E.R.; Weissman, J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2013, 2, e01179. [Google Scholar] [CrossRef]
- Li, G.W.; Oh, E.; Weissman, J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 2012, 484, 538–541. [Google Scholar] [CrossRef]
- Liu, M.J.; Wu, S.H.; Wu, J.F.; Lin, W.D.; Wu, Y.C.; Tsai, T.Y.; Tsai, H.L.; Wu, S.H. Translational landscape of photomorphogenic Arabidopsis. Plant Cell 2013, 25, 3699–3710. [Google Scholar] [CrossRef] [PubMed]
- Shalgi, R.; Hurt, J.A.; Krykbaeva, I.; Taipale, M.; Lindquist, S.; Burge, C.B. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 2013, 49, 439–452. [Google Scholar] [CrossRef]
- Stadler, M.; Artiles, K.; Pak, J.; Fire, A. Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 2012, 22, 2418–2426. [Google Scholar] [CrossRef]
- Halbeisen, R.E.; Gerber, A.P. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 2009, 7, e1000105. [Google Scholar] [CrossRef] [PubMed]
- Hettiarachchi, G.K.; Katneni, U.K.; Hunt, R.C.; Kames, J.M.; Athey, J.C.; Bar, H.; Sauna, Z.E.; McGill, J.R.; Ibla, J.C.; Kimchi-Sarfaty, C. Translational and transcriptional responses in human primary hepatocytes under hypoxia. Am. J. Physiol. Gastrointest. Liver. Physiol. 2019, 316, G720–G734. [Google Scholar] [CrossRef] [PubMed]
- Poidevin, L.; Forment, J.; Unal, D.; Ferrando, A. Transcriptome and translatome changes in germinated pollen under heat stress uncover roles of transporter genes involved in pollen tube growth. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Leushkin, E.; Liechti, A.; Ovchinnikova, S.; Mößinger, K.; Brüning, T.; Rummel, C.; Grützner, F.; Cardoso-Moreira, M.; Janich, P.; et al. Transcriptome and translatome co-evolution in mammals. Nature 2020, 588, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zeng, L.; Zhang, Z. Translatome and Transcriptome Profiling of Hypoxic-Induced Rat Cardiomyocytes. Mol. Nucleic Acids 2020, 22, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Ishibashi, Y.; Shinmyo, A.; Kanaya, S.; Kato, K. Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 448–462. [Google Scholar] [CrossRef]
- Preiss, T.; Baron-Benhamou, J.; Ansorge, W.; Hentze, M.W. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol. 2003, 10, 1039–1047. [Google Scholar] [CrossRef]
- Li, Y.F.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018, 19, 935. [Google Scholar] [CrossRef]
- Melamed, D.; Pnueli, L.; Arava, Y. Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. RNA 2008, 14, 1337–1351. [Google Scholar] [CrossRef] [PubMed]
- Gerashchenko, M.V.; Lobanov, A.V.; Gladyshev, V.N. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 2012, 109, 17394–17399. [Google Scholar] [CrossRef] [PubMed]
- Lukoszek, R.; Feist, P.; Ignatova, Z. Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-Seq. BMC Plant Biol. 2016, 16, 221. [Google Scholar] [CrossRef] [PubMed]
- Juntawong, P.; Girke, T.; Bazin, J.; Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, E203–E212. [Google Scholar] [CrossRef]
- Bazin, J.; Baerenfaller, K.; Gosai, S.J.; Gregory, B.D.; Crespi, M.; Bailey-Serres, J. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. USA 2017, 114, E10018–E10027. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Greene, G.H.; Yoo, H.; Liu, L.; Marqués, J.; Motley, J.; Dong, X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Zid, B.M.; O’Shea, E.K. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 2014, 514, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Protter, D.S.W.; Parker, R. Principles and properties of stress granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Chang, I.F.; Szick-Miranda, K.; Pan, S.; Bailey-Serres, J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005, 137, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, S.; Ben-Shem, A.; Garreau de Loubresse, N.; Jenner, L.; Yusupova, G.; Yusupov, M. One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef]
- Hill, J.R.; Morris, D.R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J. Biol. Chem. 1993, 268, 726–731. [Google Scholar] [CrossRef]
- Liang, S.; Bellato, H.M.; Lorent, J.; Lupinacci, F.C.S.; Oertlin, C.; van Hoef, V.; Andrade, V.P.; Roffe, M.; Masvidal, L.; Hajj, G.N.M.; et al. Polysome-profiling in small tissue samples. Nucleic Acids Res. 2018, 46, e3. [Google Scholar] [CrossRef] [PubMed]
- Mateju, D.; Eichenberger, B.; Voigt, F.; Eglinger, J.; Roth, G.; Chao, J.A. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 2020, 183, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Posas, F.; Chambers, J.R.; Heyman, J.A.; Hoeffler, J.P.; de Nadal, E.; Ariño, J. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000, 275, 17249–17255. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Albertyn, J.; Thevelein, J.M.; Prior, B.A.; Hohmann, S. Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae. Microbiology 1999, 145, 715–727. [Google Scholar] [CrossRef]
- Łukaszuk, E.; Ciereszko, I. Plant responses to wounding stress. In Biological Diversity—From Cell to Ecosystem; Łaska, G., Ed.; Polish BotanicalSociety—Branch in Białystok: Białystok, Poland, 2012; pp. 73–85. ISBN 978-83-62069-28-6. [Google Scholar]
- Chen, P.; Liu, Q.Z. Genome-wide characterization of the WRKY gene family in cultivated strawberry (Fragaria x ananassa Duch.) and the importance of several group III members in continuous cropping. Sci. Rep. 2019, 9, 8423. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, P.K.; Sharma, A.K.; Singh, N.K.; Sonah, H.; Deshmukh, R.; Sharma, T.R. Understanding the role of the WRKY gene family under stress conditions in Pigeonpea (Cajanus Cajan L.). Plants 2019, 8, 214. [Google Scholar] [CrossRef]
- De Maio, A. Heat shock proteins: Facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar] [CrossRef]
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Cao, Y.; Ohwatari, N.; Matsumoto, T.; Kosaka, M.; Ohtsuru, A.; Yamashita, S. TGF-β1 mediates 70-kDa heat shock protein induction due to ultraviolet irradiation in human skin fibroblasts. Pflug. Arch. 1999, 438, 239–244. [Google Scholar] [CrossRef]
- Matz, J.M.; Blake, M.J.; Tatelman, H.M.; Lavoi, K.P.; Holbrook, N.J. Characterization and regulation of cold-induced heat shock protein expression in mouse brown adipose tissue. Am. J. Physiol. 1995, 269, R38–R47. [Google Scholar] [CrossRef]
- Laplante, A.F.; Moulin, V.; Auger, F.A.; Landry, J.; Li, H.; Morrow, G.; Tanguay, R.M.; Germain, L. Expression of heat shock proteins in mouse skin during wound healing. J. Histochem. Cytochem. 1998, 46, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Marvin, M.; O’Rourke, D.; Kurihara, T.; Juliano, C.E.; Harrison, K.L.; Hutson, L.D. Developmental expression patterns of the zebrafish small heat shock proteins. Dev. Dyn. 2008, 237, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Salamanca, H.H.; Antonyak, M.A.; Cerione, R.A.; Shi, H.; Lis, J.T. Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS ONE 2014, 9, e96330. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Whitesell, L.; Rogers, A.B.; Lindquist, S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 2007, 130, 1005–1018. [Google Scholar] [CrossRef]
- Size Exclusion Chromatography: Principles and Methods. Handbooks from GE Healthcare Life Sciences. Available online: https://cdn.cytivalifesciences.com/dmm3bwsv3/AssetStream.aspx?mediaformatid=10061&destinationid=10016&assetid=11639 (accessed on 10 January 2021).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic feature. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Backman, T.W.H.; Girke, T. systemPipeR: NGS workflow and report generation environment. BMC Bioinform. 2016, 17, 388. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Liu, M.; Dong, Z. Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions. Plants 2021, 10, 304. https://doi.org/10.3390/plants10020304
Chen Y, Liu M, Dong Z. Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions. Plants. 2021; 10(2):304. https://doi.org/10.3390/plants10020304
Chicago/Turabian StyleChen, Yan, Min Liu, and Zhicheng Dong. 2021. "Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions" Plants 10, no. 2: 304. https://doi.org/10.3390/plants10020304
APA StyleChen, Y., Liu, M., & Dong, Z. (2021). Preferential Ribosome Loading on the Stress-Upregulated mRNA Pool Shapes the Selective Translation under Stress Conditions. Plants, 10(2), 304. https://doi.org/10.3390/plants10020304







