A New Controlled Release System for Propolis Polyphenols and Its Biochemical Activity for Skin Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Formulation of Controlled Release Propolis Polyphenols—CRPP System
2.1.2. Physicochemical Characterization of CRPP
2.1.3. Propolis Encapsulation Efficiency in CRPP
2.1.4. Total Phenolic Content (TPC)
2.1.5. Antioxidant Activity by 2,2-diphenylpicrylhydrazyl (DPPH)
2.1.6. In Vitro Release Studies of Propolis Polyphenols from CRPP
2.1.7. Stability of CRPP
2.2. Cell Lines and Culture
2.3. Cytotoxicity Assessments
2.3.1. Sulforhodamine B (SRB) Assay
2.3.2. Measurement of Intracellular ATP Levels
2.4. UVB Assessments
2.4.1. Single Cell Gel Electrophoresis Assay (Comet Assay)
2.4.2. Assessment of Protein Carbonyl Content
2.5. Quantitative Real Time PCR(QPCR)
2.5.1. RNA Isolation and cDNA Synthesis-NHDF Cells
2.5.2. Primer Design and RT-PCR Analysis
2.6. Human Reconstituted Skin Model (EpiDermTM EPI-200)
2.6.1. Treatment and UVB Irradiation of EpiDermTM EPI-200
2.6.2. Immunohistochemistry (IHC)
2.6.3. CRPP Incorporated in an Emulsion—Reconstructed Human Skin Model
2.7. Statistical Analysis
3. Results
3.1. Characterization of CRPP
3.2. In Vitro Release Studies
3.3. Physical Chemistry
3.4. Cytotoxicity Profile of CRPP
3.5. CRPP Protects HaCat as Well as NHDF Cells under UVB Exposure Conditions
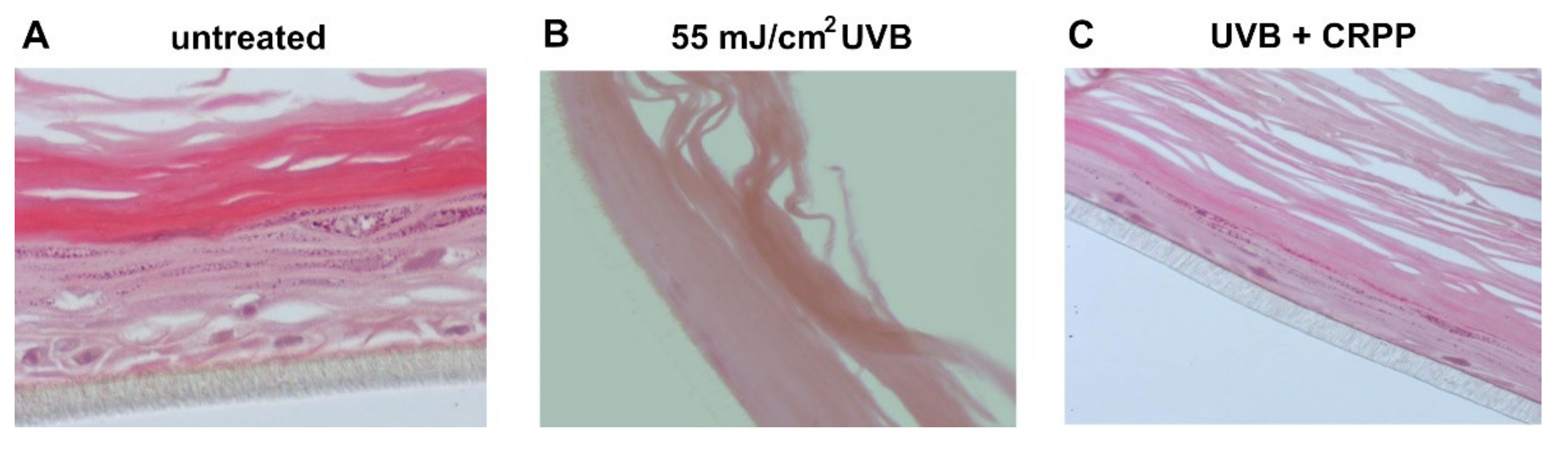
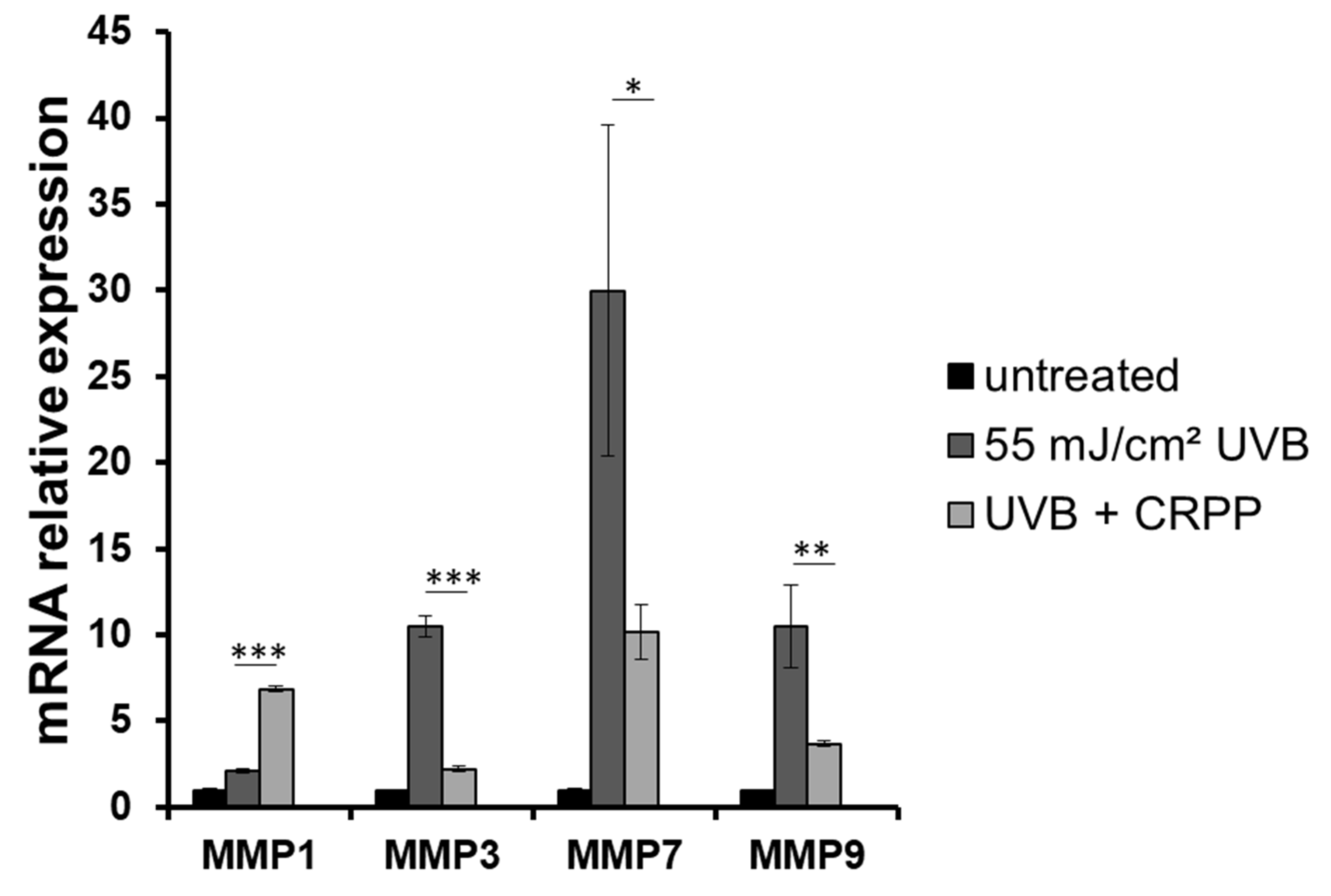
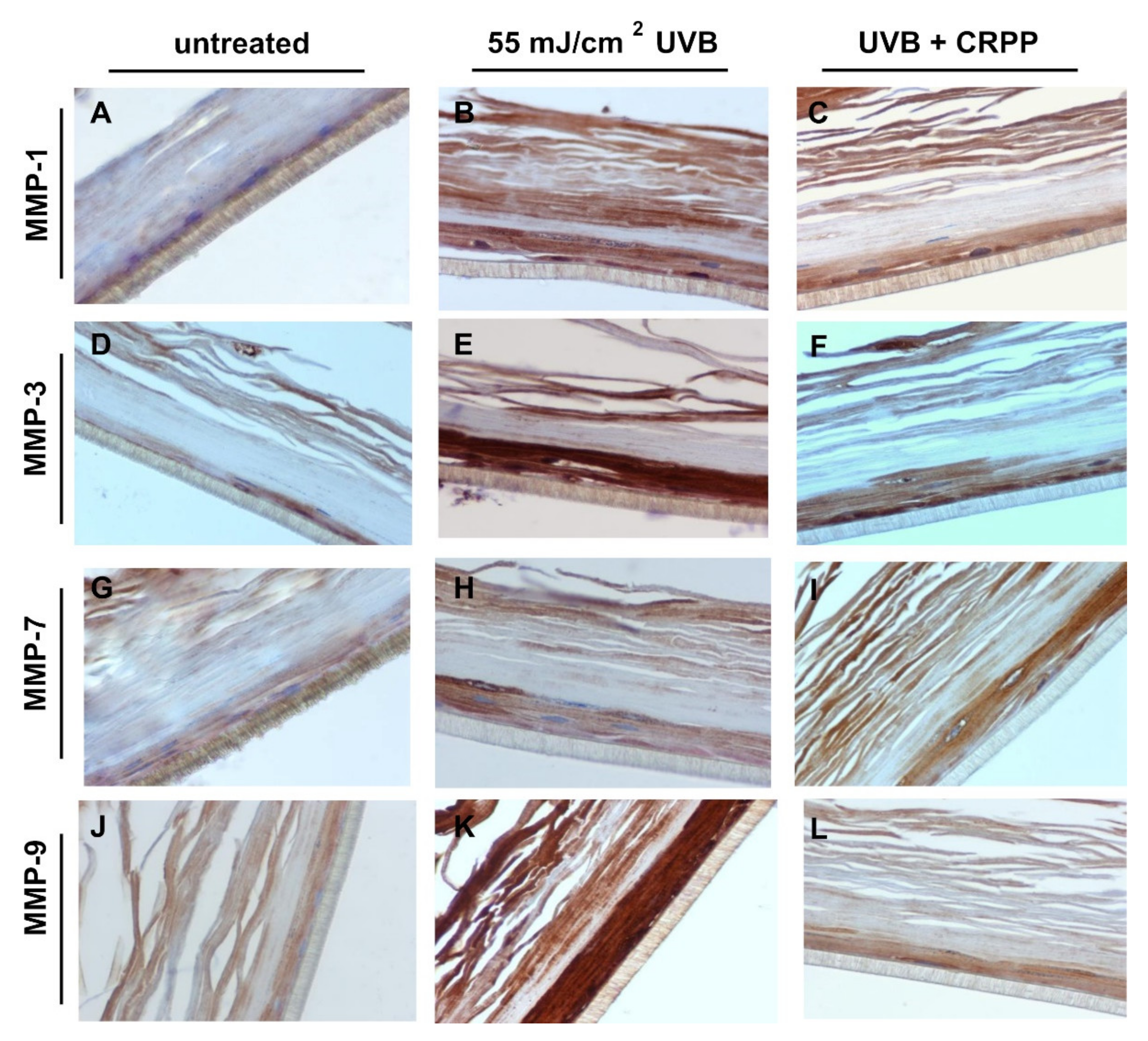
3.6. CRPP Protects Human Reconstituted Skin Model from UVB-Induced Histological Lesions and Matrix Metalloproteinases Overexpression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Münstedt, K.; Männle, H. Using Bee Products for the Prevention and Treatment of Oral Mucositis Induced by Cancer Treatment. Molecules 2019, 24, 3023. [Google Scholar] [CrossRef] [Green Version]
- Kocot, J.; Kiełczykowska, M.; Luchowska-Kocot, D.; Kurzepa, J.; Musik, I. Antioxidant Potential of Propolis, Bee Pollen, and Royal Jelly: Possible Medical Application. Oxidative Med. Cell. Longev. 2018, 2018, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Y.; Ye, Y.; Wang, X.-R.; Lin, L.-T.; Xiao, L.-Y.; Zhou, P.; Shi, G.-X.; Liu, C.-Z. Bee venom therapy: Potential mechanisms and therapeutic applications. Toxicon 2018, 148, 64–73. [Google Scholar] [CrossRef]
- Denisow, B.; Denisow-Pietrzyk, M. Biological and therapeutic properties of bee pollen: A review. J. Sci. Food Agric. 2016, 96, 4303–4309. [Google Scholar] [CrossRef]
- Przybyłek, I.; Karpiński, T.M. Antibacterial Properties of Propolis. Molecules 2019, 24, 2047. [Google Scholar] [CrossRef] [Green Version]
- Zabaiou, N.; Fouache, A.; Trousson, A.; Baron, S.; Zellagui, A.; Lahouel, M.; Lobaccaro, J.-M.A. Biological properties of propolis extracts: Something new from an ancient product. Chem. Phys. Lipids 2017, 207, 214–222. [Google Scholar] [CrossRef]
- Van Ravesteyn, L.M.; Berg, M.P.L.-V.D.; Hoogendijk, W.J.G.; Kamperman, A.M. Interventions to treat mental disorders during pregnancy: A systematic review and multiple treatment meta-analysis. PLoS ONE 2017, 12, e0173397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjum, S.I.; Ullah, A.; Khan, K.A.; Attaullah, M.; Khan, H.; Ali, H.; Bashir, M.A.; Tahir, M.; Ansari, M.J.; Ghramh, H.A.; et al. Composition and functional properties of propolis (bee glue): A review. Saudi J. Biol. Sci. 2019, 26, 1695–1703. [Google Scholar] [CrossRef] [PubMed]
- El-Guendouz, S.; Lyoussi, B.; Miguel, M.G.C. Insight on Propolis from Mediterranean Countries: Chemical Composition, Biological Activities and Application Fields. Chem. Biodivers. 2019, 16, e1900094. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, N.; Cuevas, A.; Cavalcante, M.F.; Dörr, F.A.; Saavedra, K.; Zambrano, T.; Abdalla, D.S.P.; Salazar, L.A. Polyphenols from Chilean Propolis and Pinocembrin Reduce MMP-9 Gene Expression and Activity in Activated Macrophages. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Pickard, S.; Baraitser, P.; Rymer, J.; Piper, J. Can gynaecology teaching associates provide high quality effective training for medical students in the United Kingdom? Comparative study. BMJ 2003, 327, 1389–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vukovic, N.L.; Obradović, A.D.; Vukic, M.D.; Jovanović, D.; Djurdjevic, P.M. Cytotoxic, proapoptotic and antioxidative potential of flavonoids isolated from propolis against colon (HCT-116) and breast (MDA-MB-231) cancer cell lines. Food Res. Int. 2018, 106, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Aminimoghadamfarouj, N.; Nematollahi, A. Propolis Diterpenes as a Remarkable Bio-Source for Drug Discovery Development: A Review. Int. J. Mol. Sci. 2017, 18, 1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misir, S.; Aliyazicioglu, Y.; Demir, S.; Turan, I.; Hepokur, C. Effect of Turkish Propolis on miRNA Expression, Cell Cycle, and Apoptosis in Human Breast Cancer (MCF-7) Cells. Nutr. Cancer 2019, 72, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Popova, M.; Giannopoulou, E.; Skalicka-Woźniak, K.; Graikou, K.; Widelski, J.; Bankova, V.; Kalofonos, H.; Sivolapenko, G.; Gaweł-Bęben, K.; Antosiewicz, B.; et al. Characterization and Biological Evaluation of Propolis from Poland. Molecules 2017, 22, 1159. [Google Scholar] [CrossRef]
- Kapare, H.; Lohidasan, S.; Sinnathambi, A.; Mahadik, K. Standardization, anti-carcinogenic potential and biosafety of Indian propolis. J. Ayurveda Integr. Med. 2019, 10, 81–87. [Google Scholar] [CrossRef]
- Devequi-Nunes, D.; Machado, B.A.S.; Barreto, G.D.A.; Silva, J.R.; Da Silva, D.F.; Da Rocha, J.L.C.; Brandão, H.N.; Borges, V.M.; Umsza-Guez, M.A. Chemical characterization and biological activity of six different extracts of propolis through conventional methods and supercritical extraction. PLoS ONE 2018, 13, e0207676. [Google Scholar] [CrossRef]
- Kubilienė, L.; Jekabsone, A.; Žilius, M.; Trumbeckaitė, S.; Simanaviciute, D.; Gerbutaviciene, R.; Majiene, D. Comparison of aqueous, polyethylene glycol-aqueous and ethanolic propolis extracts: Antioxidant and mitochondria modulating properties. BMC Complement. Altern. Med. 2018, 18, 165. [Google Scholar] [CrossRef]
- Cavalaro, R.I.; Da Cruz, R.G.; Dupont, S.; Bell, J.M.L.N.D.M.; Vieira, T.M.F.D.S. In vitro and in vivo antioxidant properties of bioactive compounds from green propolis obtained by ultrasound-assisted extraction. Food Chem. X 2019, 4, 100054. [Google Scholar] [CrossRef]
- Jug, M.; Karas, O.; Kosalec, I. The Influence of Extraction Parameters on Antimicrobial Activity of Propolis Extracts. Nat. Prod. Commun. 2017, 12, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Silva, I.S.D.M.; Gaspar, L.M.D.A.C.; Rocha, A.M.O.; Da Costa, L.P.; Tada, D.B.; Franceschi, E.; Padilha, F.F. Encapsulation of Red Propolis in Polymer Nanoparticles for the Destruction of Pathogenic Biofilms. AAPS PharmSciTech 2020, 21, 1–9. [Google Scholar] [CrossRef]
- Andrade, J.K.S.; Denadai, M.; Andrade, G.R.S.; Nascimento, C.D.C.; Barbosa, P.F.; Jesus, M.S.; Narain, N. Development and characterization of microencapsules containing spray dried powder obtained from Brazilian brown, green and red propolis. Food Res. Int. 2018, 109, 278–287. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, Y.; Xu, Y.; Niu, F.; Li, Z.; Ba, C.; Jin, B.; Chen, G.; Li, X. One-step assembly of zein/caseinate/alginate nanoparticles for encapsulation and improved bioaccessibility of propolis. Food Funct. 2019, 10, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin influences quorum sensing in food borne bacteria: In-vitro and in-silico evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, T.G.D.; Da Silva, P.F.; Azevedo, L.F.; Da Rocha, L.G.; Porto, I.C.C.D.M.; E Moura, T.F.A.L.; Basílio-Júnior, I.D.; Grillo, L.A.M.; Dornelas, C.B.; Fonseca, E.J.D.S.; et al. Polymeric Nanoparticles of Brazilian Red Propolis Extract: Preparation, Characterization, Antioxidant and Leishmanicidal Activity. Nanoscale Res. Lett. 2016, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aytekin, A.A.; Tanrıverdi, S.T.; Köse, F.A.; Kart, D.; Eroğlu, I.; Özer, Ö. Propolis loaded liposomes: Evaluation of antimicrobial and antioxidant activities. J. Liposome Res. 2019, 30, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, D.; Hu, Y.; Huang, Y.; Yu, Y.; Wang, D. The Immunological Enhancement Activity of Propolis Flavonoids LiposomeIn VitroandIn Vivo. Evidence Based Complement. Altern. Med. 2014, 2014, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arafa, M.G.; Ghalwash, D.; El-Kersh, D.M.; Elmazar, M.M. Propolis-based niosomes as oromuco-adhesive films: A randomized clinical trial of a therapeutic drug delivery platform for the treatment of oral recurrent aphthous ulcers. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Sharaf, S.; El-Naggar, M.E. Wound dressing properties of cationized cotton fabric treated with carrageenan/cyclodextrin hydrogel loaded with honey bee propolis extract. Int. J. Biol. Macromol. 2019, 133, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulos, N.; Konteles, S.; Mourtzinos, I.; Troullidou, E.; Chiou, A.; Karathanos, V.T. Encapsulation of complex extracts inβ-cyclodextrin: An application to propolis ethanolic extract. J. Microencapsul. 2009, 26, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Bordonaro, M.; Lazarova, D. Hypothesis: Induction of biomarkers for detection of colonic neoplasms. J. Cancer 2018, 9, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, R.; Nigam, N.; Bhargava, P.; Dhanjal, J.K.; Goyal, S.; Grover, A.; Sundar, D.; Ishida, Y.; Terao, K.; Kaul, S.C. Molecular Characterization and Enhancement of Anticancer Activity of Caffeic Acid Phenethyl Ester by γ Cyclodextrin. J. Cancer 2016, 7, 1755–1771. [Google Scholar] [CrossRef] [Green Version]
- Vasilaki, A.; Hatzikamari, M.; Stagkos-Georgiadis, A.; Goula, A.M.; Mourtzinos, I. A natural approach in food preservation: Propolis extract as sorbate alternative in non-carbonated beverage. Food Chem. 2019, 298, 125080. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Gao, R.; Shah, N.; Bhargava, P.; Furune, T.; Kaul, S.C.; Terao, K.; Wadhwa, R. Anticancer Activity in Honeybee Propolis: Functional Insights to the Role of Caffeic Acid Phenethyl Ester and Its Complex With γ-Cyclodextrin. Integr. Cancer Ther. 2018, 17, 867–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rimbach, G.; Fischer, A.; Schloesser, A.; Jerz, G.; Ikuta, N.; Ishida, Y.; Matsuzawa, R.; Matsugo, S.; Huebbe, P.; Terao, K. Anti-Inflammatory Properties of Brazilian Green Propolis Encapsulated in a γ-Cyclodextrin Complex in Mice Fed a Western-Type Diet. Int. J. Mol. Sci. 2017, 18, 1141. [Google Scholar] [CrossRef] [Green Version]
- Zappacosta, R.; Cornelio, B.; Pilato, S.; Siani, G.; Estour, F.; Aschi, M.; Fontana, A. Effect of the Incorporation of Functionalized Cyclodextrins in the Liposomal Bilayer. Molecules 2019, 24, 1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, E.-Y.; Chen, Y.-S.; Li, Y.-S.; Chen, S.-R.; Lee, C.-H.; Huang, M.-H.; Chuang, H.-M.; Harn, H.-J.; Yang, H.-H.; Lin, S.-Z.; et al. Liposome Consolidated with Cyclodextrin Provides Prolonged Drug Retention Resulting in Increased Drug Bioavailability in Brain. Int. J. Mol. Sci. 2020, 21, 4408. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ran, L.; Liu, F.; Hou, R.; Zhao, W.; Li, Y.; Wang, C.; Dong, J. Preparation and Characterisation of Polyphenol-HP-β-Cyclodextrin Inclusion Complex that Protects Lamb Tripe Protein against Oxidation. Molecules 2019, 24, 4487. [Google Scholar] [CrossRef] [Green Version]
- Cutrignelli, A.; Lopedota, A.; Denora, N.; Iacobazzi, R.M.; Fanizza, E.; Laquintana, V.; Perrone, M.; Maggi, V.; Franco, M. A New Complex of Curcumin with Sulfobutylether-β-Cyclodextrin: Characterization Studies and In Vitro Evaluation of Cytotoxic and Antioxidant Activity on HepG-2 Cells. J. Pharm. Sci. 2014, 103, 3932–3940. [Google Scholar] [CrossRef]
- Panahi, Y.; Farshbaf, M.; Mohammadhosseini, M.; Mirahadi, M.; Khalilov, R.; Saghfi, S.; Akbarzadeh, A. Recent advances on liposomal nanoparticles: Synthesis, characterization and biomedical applications. Artif. Cells Nanomed. Biotechnol. 2017, 45, 788–799. [Google Scholar] [CrossRef] [Green Version]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Du, C.; Guo, N.; Teng, Y.; Meng, X.; Sun, H.; Li, S.; Yu, P.; Galons, H. Composition design and medical application of liposomes. Eur. J. Med. Chem. 2019, 164, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Karapetsas, A.; Voulgaridou, G.-P.; Konialis, M.; Tsochantaridis, I.; Kynigopoulos, S.; Lambropoulou, M.; Stavropoulou, M.-I.; Stathopoulou, K.; Aligiannis, N.; Bozidis, P.; et al. Propolis Extracts Inhibit UV-Induced Photodamage in Human Experimental In Vitro Skin Models. Antioxidants 2019, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Gardikis, K.; Hatziantoniou, S.; Signorelli, M.; Pusceddu, M.; Micha-Screttas, M.; Schiraldi, A.; Demetzos, C.; Fessas, D. Thermodynamic and structural characterization of Liposomal-Locked in-Dendrimers as drug carriers. Colloids Surf. B Biointerfaces 2010, 81, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.G.; Syed, S.; Marshall, K.; Hosseinidoust, Z. Liposomal Nanovesicles for Efficient Encapsulation of Staphylococcal Antibiotics. ACS Omega 2019, 4, 10866–10876. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Arnous, A.; Makris, D.P.; Kefalas, P. Correlation of Pigment and Flavanol Content with Antioxidant Properties in Selected Aged Regional Wines from Greece. J. Food Compos. Anal. 2002, 15, 655–665. [Google Scholar] [CrossRef]
- Giampieri, F.; Alvarez-Suarez, J.M.; Mazzoni, L.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Gonzàlez-Paramàs, A.M.; Santos-Buelga, C.; Quiles, J.L.; Bompadre, S.; Mezzetti, B.; et al. Polyphenol-Rich Strawberry Extract Protects Human Dermal Fibroblasts against Hydrogen Peroxide Oxidative Damage and Improves Mitochondrial Functionality. Molecules 2014, 19, 7798–7816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ramakersab, C.; Ruijter, J.M.; Deprez, R.H.; Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Weber, D.; Davies, M.J.; Grune, T. Determination of protein carbonyls in plasma, cell extracts, tissue homogenates, isolated proteins: Focus on sample preparation and derivatization conditions. Redox Biol. 2015, 5, 367–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Milzani, A.; Colombo, R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 2003, 329, 23–38. [Google Scholar] [CrossRef]
- Karapetsas, A.; Voulgaridou, G.-P.; Iliadi, D.; Tsochantaridis, I.; Michail, P.; Kynigopoulos, S.; Lambropoulou, M.; Stavropoulou, M.-I.; Stathopoulou, K.; Karabournioti, S.; et al. Honey Extracts Exhibit Cytoprotective Properties against UVB-Induced Photodamage in Human Experimental Skin Models. Antioxidants 2020, 9, 566. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Hepburn, H.; Li, Y.; Chen, M.; Radloff, S.; Daya, S. Effects of ethanol and water extracts of propolis (bee glue) on acute inflammatory animal models. J. Ethnopharmacol. 2005, 100, 276–283. [Google Scholar] [CrossRef]
- Crouch, S.; Kozlowski, R.; Slater, K.; Fletcher, J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J. Immunol. Methods 1993, 160, 81–88. [Google Scholar] [CrossRef]
- Deters, A.M.; Schröder, K.R.; Hensel, A. Kiwi fruit (Actinidia chinensis L.) polysaccharides exert stimulating effects on cell proliferation via enhanced growth factor receptors, energy production, and collagen synthesis of human keratinocytes, fibroblasts, and skin equivalents. J. Cell. Physiol. 2004, 202, 717–722. [Google Scholar] [CrossRef]
- Fehsel, K.; Kolb-Bachofen, V.; Kolb, H. Analysis of TNFα-induced DNA strand breaks at the single cell level. Am. J. Pathol. 1991, 139, 251–254. [Google Scholar]
- Alikhani, M.; Alikhani, Z.; Raptis, M.; Graves, D.T. TNF-? in vivo stimulates apoptosis in fibroblasts through caspase-8 activation and modulates the expression of pro-apoptotic genes. J. Cell. Physiol. 2004, 201, 341–348. [Google Scholar] [CrossRef]
- Na, J.; Bak, D.; Im, S.I.; Choi, H.; Hwang, J.H.; Kong, S.Y.; No, Y.A.; Lee, Y.; Kim, B.J. Anti-apoptotic effects of glycosaminoglycans via inhibition of ERK/AP-1 signaling in TNF-α-stimulated human dermal fibroblasts. Int. J. Mol. Med. 2018, 41, 3090–3098. [Google Scholar] [CrossRef] [PubMed]
- Moens, S.; Goveia, J.; Stapor, P.C.; Cantelmo, A.R.; Carmeliet, P. The multifaceted activity of VEGF in angiogenesis—Implications for therapy responses. Cytokine Growth Factor Rev. 2014, 25, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Badiu, D.; Vasile, M.; Teren, O. Regulation of wound healing by growth factors and cytokines. Wound Health Process. Phases Promot. 2011, 3, 73–93. [Google Scholar]
- Letsiou, S.; Kalliampakou, K.; Gardikis, K.; Mantecon, L.; Infante, C.; Chatzikonstantinou, M.; Labrou, N.E.; Flemetakis, E. Skin Protective Effects of Nannochloropsis gaditana Extract on H2O2-Stressed Human Dermal Fibroblasts. Front. Mar. Sci. 2017, 4, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Morgan, C.; Nigam, Y. Naturally derived factors and their role in the promotion of angiogenesis for the healing of chronic wounds. Angiogenesis 2013, 16, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Kant, V.; Gopal, A.; Kumar, D.; Pathak, N.N.; Ram, M.; Jangir, B.L.; Tandan, S.K.; Kumar, D. Curcumin-induced angiogenesis hastens wound healing in diabetic rats. J. Surg. Res. 2015, 193, 978–988. [Google Scholar] [CrossRef]
- Hara-Chikuma, M.; Verkman, A. Roles of Aquaporin-3 in the Epidermis. J. Investig. Dermatol. 2008, 128, 2145–2151. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Sun, Y.; Healey, S.; Bi, Z.; Hu, G.; Wan, S.; Kouttab, N.; Chu, W.; Wan, Y. EGFR-mediated expression of aquaporin-3 is involved in human skin fibroblast migration. Biochem. J. 2006, 400, 225–234. [Google Scholar] [CrossRef]
- Junttila, I.S. Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, M.; Werfel, T. Innate immunity, allergy and atopic dermatitis. Curr. Opin. Allergy Clin. Immunol. 2010, 10, 463–468. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H. Integrin signalling and function in immune cells. Immunology 2012, 135, 268–275. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Chen, B.; Humeres, C.; Alex, L.; Hanna, A.; Frangogiannis, N.G. The role of Smad2 and Smad3 in regulating homeostatic functions of fibroblasts in vitro and in adult mice. Biochim. Biophys. Acta (BBA) Bioenerg. 2020, 1867, 118703. [Google Scholar] [CrossRef]





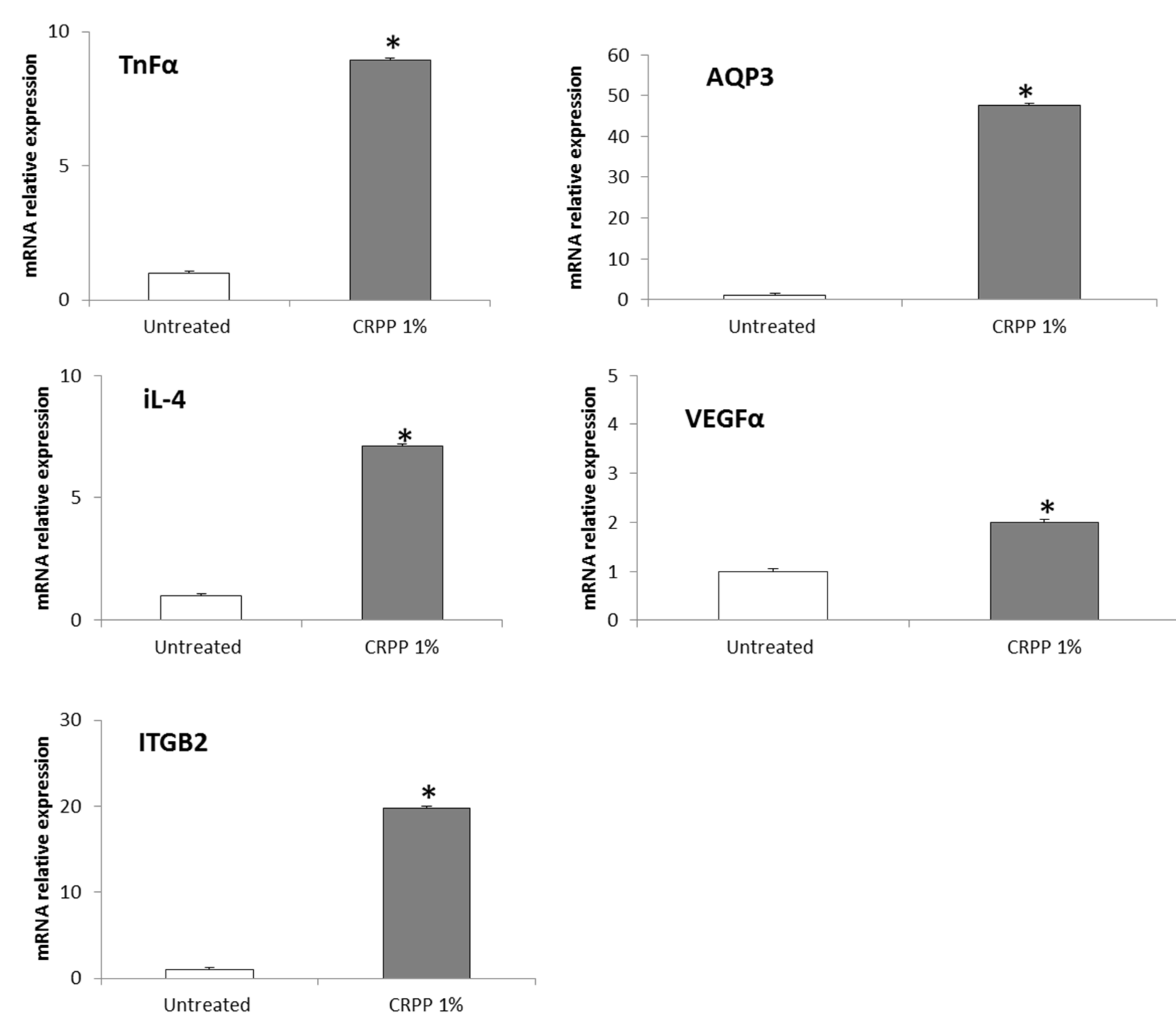

| Gene Symbol | Gene Name | Primmer F (5’-3’) | Primmer R (5’-3’) |
|---|---|---|---|
| ACTB | beta actin | CTGTCCACCTTCCAGCAGATGT | AGCATTTGCGGTGGACGAT |
| TNFa | Tumor necrosis factor alpha | GCCCTGGTATGAGCCCATCTAT | ATCCCAAAGTAGACCTGCCCA |
| AQP3 | Aquaporin 3 | TGTACCAGCTGATGATCGGCT | GATCTGCTCCTTGTGCTTCACA |
| VEGFα | vascular endothelial growth factor A | CAGACGTGTAAATGTTCCTGCA | ACGTTCGTTTAACTCAAGCTGC |
| IL4 | Interleukin 4 | TTCCTGAAACGGCTCGACA | CGTACTCTGGTTGGCTTCCTTC |
| ITGB2 | Integrin subunit beta 2 | TGCATCCAGGAGCAGTCGTTT | GGAAGAACCTGCACGGTCACTA |
| A/A | INCI (EU) | % (w/w) |
|---|---|---|
| 1 | Aqua | 71.97 |
| 2 | Neopentyl Glycol Diheptanoate | 4.08 |
| 3 | Dicaprylyl Ether | 3.50 |
| 4 | Cetearyl Alcohol | 3.13 |
| 5 | Glycerin | 3.00 |
| 6 | Cocoglycerides | 3.00 |
| 7 | Glyceryl Stearate | 2.00 |
| 8 | Propylene glycol | 1.65 |
| 9 | PEG-100 Stearate | 1.20 |
| 10 | CRPP | 1.00 |
| 11 | PEG-20 Stearate | 0.67 |
| 12 | Phenoxyethanol | 0.55 |
| 13 | Tocopheryl Acetate | 0.50 |
| 14 | Panthenol | 0.50 |
| 15 | Sorbitan Stearate | 0.50 |
| 16 | Caprylic/Capric Triglyceride | 0.45 |
| 17 | Caprylyl Glycol | 0.46 |
| 18 | Vitis vinifera leaf extract | 0.35 |
| 19 | Ethylhexylglycerin | 0.30 |
| 20 | Carbomer | 0.30 |
| 21 | Allantoin | 0.20 |
| 22 | Bisabolol | 0.20 |
| 23 | Sodium Hydroxide | 0.20 |
| 24 | Disodium EDTA | 0.15 |
| 25 | Tocopherol | 0.15 |
| Total | 100 |
| System | z-Average Diameter (nm) | SD | PI | SD | ζ-pot (mV) | SD |
|---|---|---|---|---|---|---|
| CRPP day 1 | 255 | 36 | 0.355 | 0.051 | −21 | 5.0 |
| CRPP 25 °C day 30 | 299 | 28 | 0.401 | 0.012 | −16 | 2.2 |
| CRPP 6 °C day 30 | 249 | 51 | 0.286 | 0.036 | −21 | 3.0 |
| CRPP 38 °C day 30 | 402 | 44 | 0.216 | 0.038 | −19 | 3.7 |
| Week | Parameters | TPC (μg GA/mL) | DPPH (mM Trolox Equivalent) | Density (g/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T(0) | 100.9 | ± | 0.7 | 33.67 | ± | 0.16 | 1.0502 | ± | 0.0006 | |
| 4th week | RT | 88.2 b,d,e | ± | 1.7 | 29.11 d,e,f | ± | 0.34 | 1.0513 | ± | 0.0004 |
| 6 °C | 88.9 c,d,e | ± | 1.7 | 29.38 a,b,c,d | ± | 0.49 | 1.0506 | ± | 0.0004 | |
| 38 °C | 80.7 h | ± | 1.2 | 30.22 a | ± | 0.23 | 1.0514 | ± | 0.0003 | |
| 8th week | RT | 85.2 a,d,f,g | ± | 1.2 | 27.69 h | ± | 0.58 | 1.0509 | ± | 0.0012 |
| 6 °C | 88.9 a,b,c | ± | 0.34 | 28.28 b,e,g,h | ± | 0.67 | 1.0489 | ± | 0.0013 | |
| 38 °C | 79.4 h | ± | 2.2 | 28.84 c,f,g | ± | 0.79 | 1.0513 | ± | 0.0012 | |
| 24th week | RT | 70.9 | ± | 0.7 | 22.13 | ± | 0.72 | 1.0505 | ± | 0.0005 |
| 6 °C | 84.5 g | ± | 3.9 | 23.90 | ± | 0.23 | 1.0497 | ± | 0.0012 | |
| 38 °C | 61.6 | ± | 0.35 | 25.62 | ± | 0.47 | 1.0520 | ± | 0.0013 | |
| Week | Parameters | Aspect | Color | Odor | pH (25 °C) | Refractive Index (Brix %) |
|---|---|---|---|---|---|---|
| T(0) | Liquid | Dark Yellow Turbid | Characteristic | 6.08 | 37.38 | |
| 1st Week | RT | N | N | N | 5.99 | 37.29 |
| 6 °C | N | N | N | 6.01 | 37.01 | |
| 38 °C | N | N | N | 5.96 | 37.35 | |
| 2nd Week | RT | N | N | N | 5.66 | 37.71 |
| 6 °C | N | N | N | 6.06 | 37.77 | |
| 38 °C | N | N | N | 5.66 | 38.10 | |
| 3rd Week | RT | N | N | N | 5.88 | 38.00 |
| 6 °C | N | N | N | 6.09 | 37.58 | |
| 38 °C | N | N | N | 5.79 | 37.81 | |
| 4th Week | RT | N | N | N | 5.97 | 37.53 |
| 6 °C | N | N | N | 6.08 | 37.58 | |
| 38 °C | N | M | N | 5.80 | 37.69 | |
| 8th Week | RT | N | N | N | 5.80 | 38.99 |
| 6 °C | N | N | N | 6.05 | 37.50 | |
| 38 °C | N | M | N | 5.70 | 38.01 | |
| 12th Week | RT | N | N | N | 5.72 | 37.51 |
| 6 °C | N | N | N | 5.90 | 37.53 | |
| 38 °C | N | M | N | 5.55 | 37.85 | |
| 24th Week | RT | N | N | N | 5.67 | 38.08 |
| 6 °C | N | N | N | 5.90 | 37.97 | |
| 38 °C | N | M | N | 5.56 | 47.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spanidi, E.; Karapetsas, A.; Voulgaridou, G.-P.; Letsiou, S.; Aligiannis, N.; Tsochantaridis, I.; Kynigopoulos, S.; Lambropoulou, M.; Mourtzinos, I.; Pappa, A.; et al. A New Controlled Release System for Propolis Polyphenols and Its Biochemical Activity for Skin Applications. Plants 2021, 10, 420. https://doi.org/10.3390/plants10020420
Spanidi E, Karapetsas A, Voulgaridou G-P, Letsiou S, Aligiannis N, Tsochantaridis I, Kynigopoulos S, Lambropoulou M, Mourtzinos I, Pappa A, et al. A New Controlled Release System for Propolis Polyphenols and Its Biochemical Activity for Skin Applications. Plants. 2021; 10(2):420. https://doi.org/10.3390/plants10020420
Chicago/Turabian StyleSpanidi, Eleni, Athanasios Karapetsas, Georgia-Persephoni Voulgaridou, Sophia Letsiou, Nektarios Aligiannis, Ilias Tsochantaridis, Spyridon Kynigopoulos, Maria Lambropoulou, Ioannis Mourtzinos, Aglaia Pappa, and et al. 2021. "A New Controlled Release System for Propolis Polyphenols and Its Biochemical Activity for Skin Applications" Plants 10, no. 2: 420. https://doi.org/10.3390/plants10020420
APA StyleSpanidi, E., Karapetsas, A., Voulgaridou, G.-P., Letsiou, S., Aligiannis, N., Tsochantaridis, I., Kynigopoulos, S., Lambropoulou, M., Mourtzinos, I., Pappa, A., & Gardikis, K. (2021). A New Controlled Release System for Propolis Polyphenols and Its Biochemical Activity for Skin Applications. Plants, 10(2), 420. https://doi.org/10.3390/plants10020420











