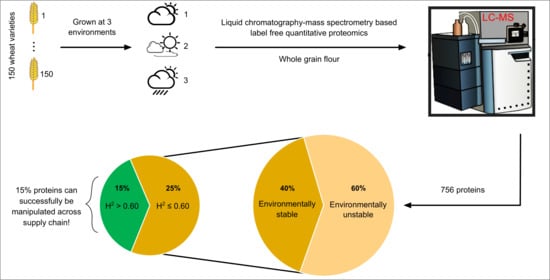
Characterization of 150 Wheat Cultivars by LC-MS-Based Label-Free Quantitative Proteomics Unravels Possibilities to Design Wheat Better for Baking Quality and Human Health
Abstract
1. Introduction
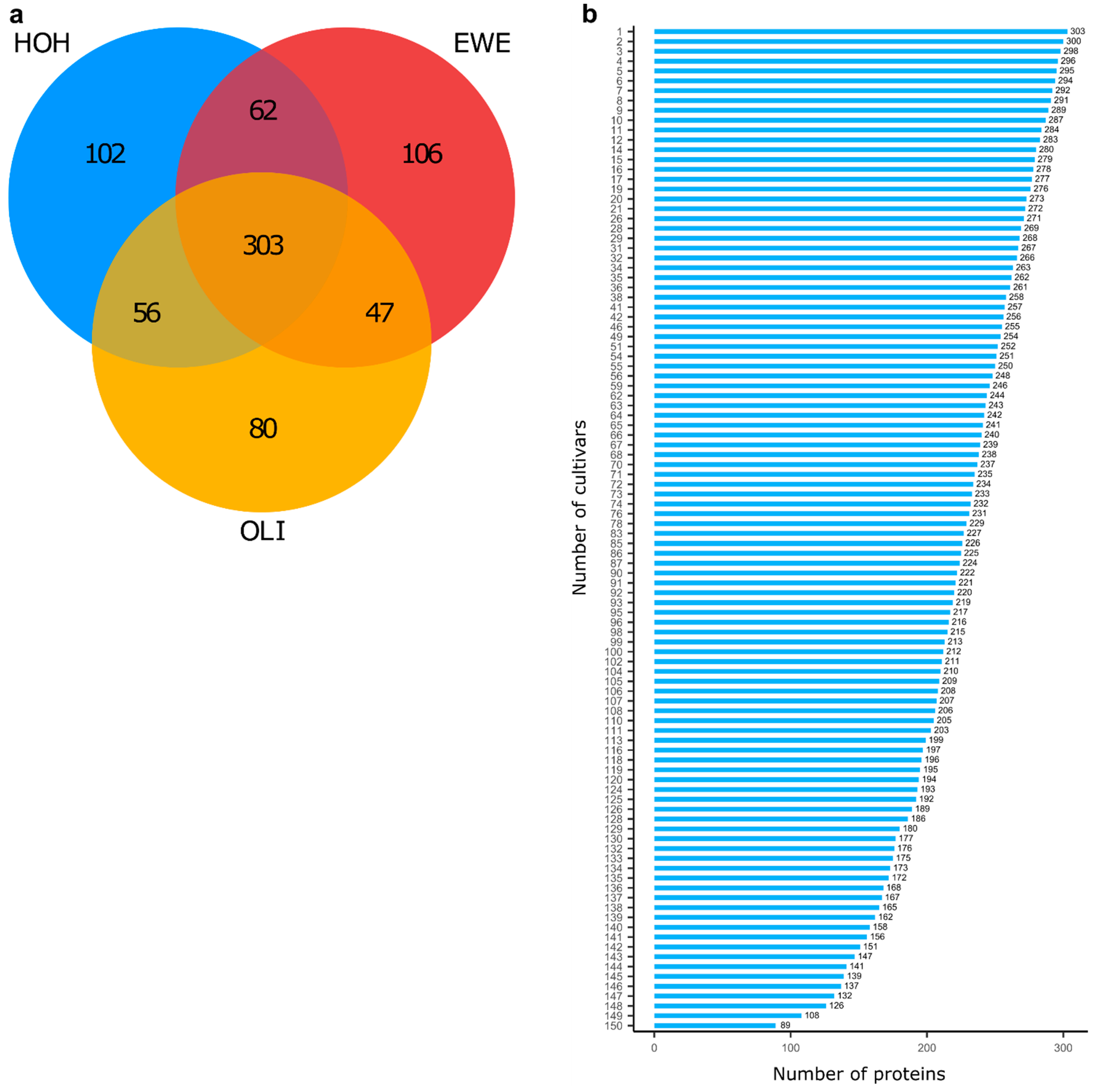
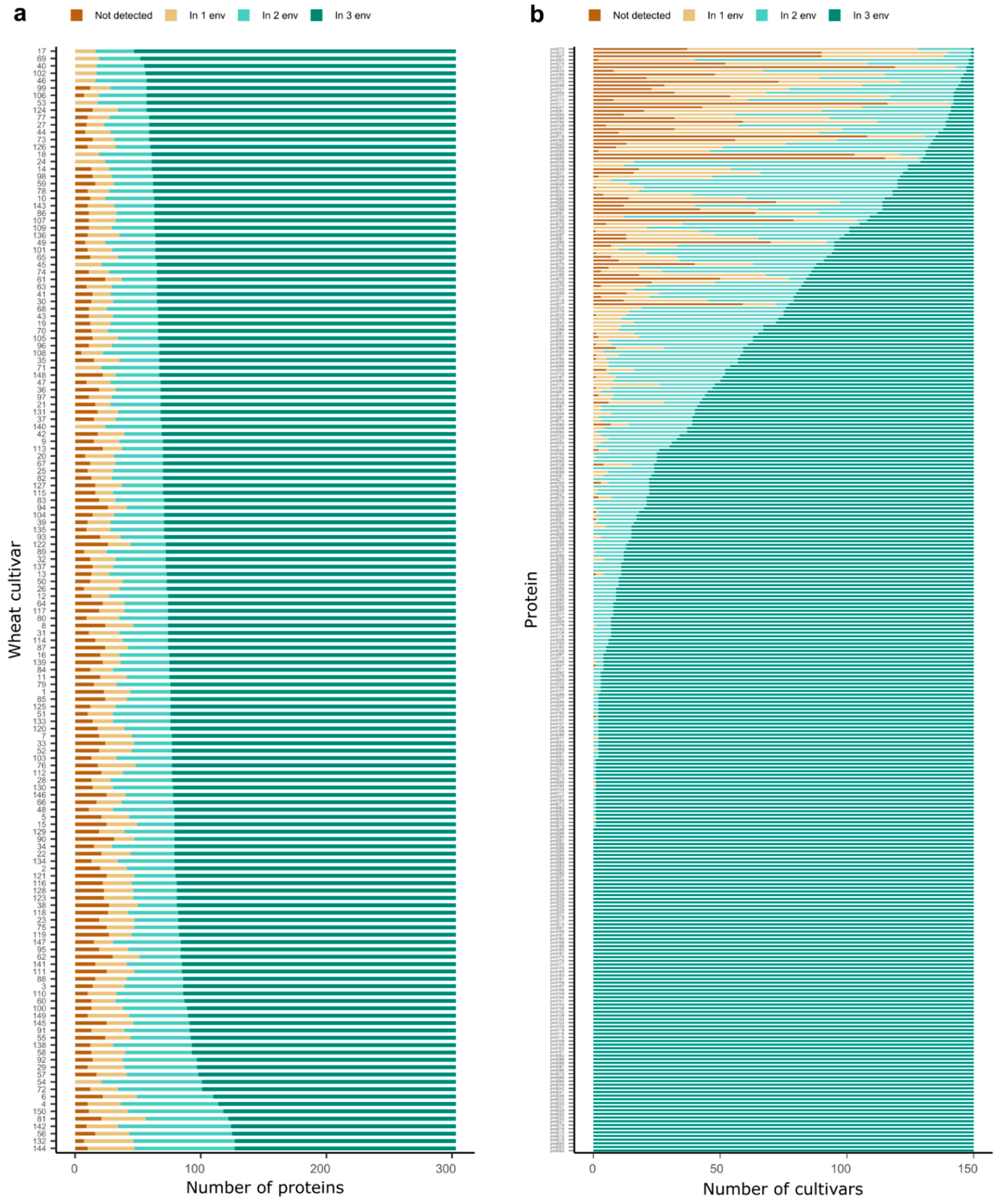
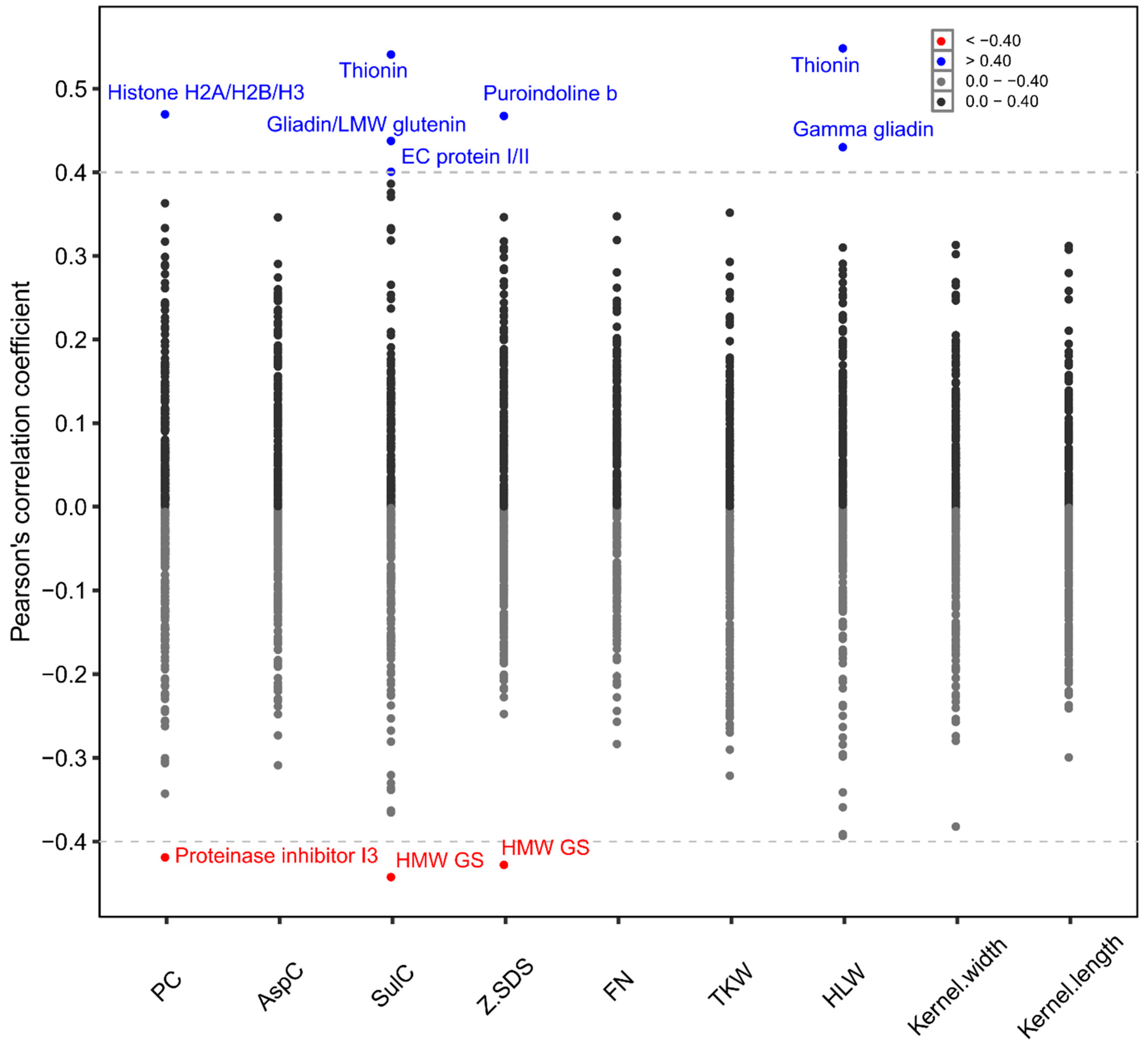
2. Results
3. Discussion
3.1. Complex Interaction between Proteins, Cultivars and Environment
3.2. Breeding for the Supply Chain
3.3. Application of New “Omics” Enables the Design of Better and Healthier Wheat
4. Materials and Methods
4.1. Plant Material and Field Trials
4.2. Quantitative Proteomic Analyses
4.3. Phenotypic Data Analysis
Detection of Temporal Trend
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Würschum, T.; Leiser, W.L.; Kazman, E.; Longin, C.F.H. Genetic control of protein content and sedimentation volume in European winter wheat cultivars. Theor. Appl. Genet. 2016, 129, 1685–1696. [Google Scholar] [CrossRef]
- Wyatt, E.C.; Bushong, J.T.; Macnack, N.E.; Mullock, J.L.; Taylor, R.; Raun, W.R. Influence of Droplet Size of Foliar-Applied Nitrogen on Grain Protein Content of Hard Red Winter Wheat. Crop. Forage Turfgrass Manag. 2017, 3, 1–10. [Google Scholar] [CrossRef]
- Johansson, E.; Branlard, G.; Cuniberti, M.; Flagella, Z.; Hüsken, A.; Nurit, E.; Peña, R.J.; Sissons, M.; Vazquez, D. Genotypic and Environmental Effects on Wheat Technological and Nutritional Quality. In Wheat Quality for Improving Processing and Human Health, 1st ed.; Igrejas, G., Ikeda, T.M., Guzmán, C., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 171–204. [Google Scholar]
- Yan, S.; Wu, Y.; Fan, J.; Zhang, F.; Zheng, J.; Qiang, S.; Guo, J.; Xiang, Y.; Zou, H.; Wu, L. Dynamic change and accumulation of grain macronutrient (N, P and K) concentrations in winter wheat under different drip fertigation regimes. Field Crop. Res. 2020, 250, 107767. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef]
- Shewry, P.R.; Tatham, A.S.; Barro, F.; Barcelo, P.; Lazzeri, P. Biotechnology of Breadmaking: Unraveling and Manipulating the Multi-Protein Gluten Complex. Nat. Biotechnol. 1995, 13, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Veraverbeke, W.S.; Delcour, J.A. Wheat Protein Composition and Properties of Wheat Glutenin in Relation to Breadmaking Functionality. Crit. Rev. Food Sci. Nutr. 2002, 42, 179–208. [Google Scholar] [CrossRef] [PubMed]
- Schuppan, D.; Junker, Y.; Barisani, D. Celiac Disease: From Pathogenesis to Novel Therapies. Gastroenterology 2009, 137, 1912–1933. [Google Scholar] [CrossRef] [PubMed]
- Sollid, L.M.; Tye-Din, J.A.; Qiao, S.-W.; Anderson, R.P.; Gianfrani, C.; Koning, F. Update 2020: Nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 2019, 72, 85–88. [Google Scholar] [CrossRef]
- Junker, Y.; Zeissig, S.; Kim, S.-J.; Barisani, D.; Wieser, H.; Leffler, D.A.; Zevallos, V.; Libermann, T.A.; Dillon, S.; Freitag, T.L.; et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 2012, 209, 2395–2408. [Google Scholar] [CrossRef]
- Schuppan, D.; Zevallos, V. Wheat Amylase Trypsin Inhibitors as Nutritional Activators of Innate Immunity. Dig. Dis. 2015, 33, 260–263. [Google Scholar] [CrossRef]
- Zevallos, V.F.; Raker, V.; Tenzer, S.; Jimenez-Calvente, C.; Ashfaq-Khan, M.; Rüssel, N.; Pickert, G.; Schild, H.; Steinbrink, K.; Schuppan, D. Nutritional Wheat Amylase-Trypsin Inhibitors Promote Intestinal Inflammation via Activation of Myeloid Cells. Gastroenterology 2017, 152, 1100–1113.e12. [Google Scholar] [CrossRef]
- Ashfaq-Khan, M.; Aslam, M.; Qureshi, M.A.; Senkowski, M.S.; Yen-Weng, S.; Strand, S.; Kim, Y.O.; Pickert, G.; Schattenberg, J.M.; Schuppan, D. Dietary wheat amylase trypsin inhibitors promote features of murine non-alcoholic fatty liver disease. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Bellinghausen, I.; Weigmann, B.; Zevallos, V.; Maxeiner, J.; Reißig, S.; Waisman, A.; Schuppan, D.; Saloga, J. Wheat amylase-trypsin inhibitors exacerbate intestinal and airway allergic immune responses in humanized mice. J. Allergy Clin. Immunol. 2019, 143, 201–212.e4. [Google Scholar] [CrossRef]
- Zevallos, V.F.; Raker, V.K.; Maxeiner, J.; Scholtes, P.; Steinbrink, K.; Schuppan, D. Dietary wheat amylase trypsin inhibitors exacerbate murine allergic airway inflammation. Eur. J. Nutr. 2018, 58, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, G.; Quirce, S.; Diaz-Perales, A. Wheat allergens associated with Baker’s asthma. J. Investig. Allergol. Clin. Immunol. 2011, 21, 81–92. [Google Scholar] [PubMed]
- Mameri, H.; Denery-Papini, S.; Pietri, M.; Tranquet, O.; Larré, C.; Drouet, M.; Paty, E.; Jonathan, A.-M.; Beaudouin, E.; Moneret-Vautrin, D.-A.; et al. Molecular and immunological characterization of wheat Serpin (Tri a 33). Mol. Nutr. Food Res. 2012, 56, 1874–1883. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Sapone, A.; Zevallos, V.; Schuppan, D. Nonceliac Gluten Sensitivity. Gastroenterology 2015, 148, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Juhász, A.; Belova, T.; Florides, C.G.; Maulis, C.; Fischer, I.; Gell, G.; Birinyi, Z.; Ong, J.; Keeble-Gagnère, G.; Maharajan, A.; et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 2018, 4, eaar8602. [Google Scholar] [CrossRef] [PubMed]
- Fritscher-Ravens, A.; Schuppan, D.; Ellrichmann, M.; Schoch, S.; Röcken, C.; Brasch, J.; Bethge, J.; Böttner, M.; Klose, J.; Milla, P.J. Confocal Endomicroscopy Shows Food-Associated Changes in the Intestinal Mucosa of Patients With Irritable Bowel Syndrome. Gastroenterology 2014, 147, 1012–1020.e4. [Google Scholar] [CrossRef] [PubMed]
- Fritscher-Ravens, A.; Pflaum, T.; Mösinger, M.; Ruchay, Z.; Röcken, C.; Milla, P.J.; Das, M.; Böttner, M.; Wedel, T.; Schuppan, D. Many Patients with Irritable Bowel Syndrome Have Atypical Food Allergies Not Associated with Immunoglobulin E. Gastroenterology 2019, 157, 109–118.e5. [Google Scholar] [CrossRef]
- EL Hassouni, K.; Sielaff, M.; Leiser, W.; Würschum, T.; Schuppan, D.; Tenzer, S.; Longin, C.F.H. Genetic Architecture Underlying the Expression of Eight Alpha-Amylase Trypsin Inhibitors as Potential Triggers of Wheat Sensitivity and Baker’s Asthma. Status (Unpublished; Manuscript in Preparation).
- Geisslitz, S.; Longin, C.F.H.; Koehler, P.; Scherf, K.A. Comparative quantitative LC–MS/MS analysis of 13 amylase/trypsin inhibitors in ancient and modern Triticum species. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Pronin, D.; Börner, A.; Scherf, K.A. Old and modern wheat (Triticum aestivum L.) cultivars and their potential to elicit celiac disease. Food Chem. 2021, 339, 127952. [Google Scholar] [CrossRef] [PubMed]
- Payne, P.I.; Corfield, K.G. Subunit composition of wheat glutenin proteins, isolated by gel filtration in a dissociating medium. Planta 1979, 145, 83–88. [Google Scholar] [CrossRef]
- Jackson, E.A.; Holt, L.M.; Payne, P.I. Characterisation of high molecular weight gliadin and low-molecular-weight glutenin subunits of wheat endosperm by two-dimensional electrophoresis and the chromosomal localisation of their controlling genes. Theor. Appl. Genet. 1983, 66, 29–37. [Google Scholar] [CrossRef]
- Pronin, D.; Börner, A.; Weber, H.; Scherf, K.A. Wheat (Triticum aestivum L.) Breeding from 1891 to 2010 Contributed to Increasing Yield and Glutenin Contents but Decreasing Protein and Gliadin Contents. J. Agric. Food Chem. 2020, 68, 13247–13256. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Hebert, A.S.; Richards, A.L.; Bailey, D.J.; Ulbrich, A.; Coughlin, E.E.; Westphall, M.S.; Coon, J.J. The One Hour Yeast Proteome. Mol. Cell. Proteom. 2014, 13, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; DeLorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Afzal, M.; Pfannstiel, J.; Zimmermann, J.; Bischoff, S.C.; Würschum, T.; Longin, C.F.H. High-resolution proteomics reveals differences in the proteome of spelt and bread wheat flour representing targets for research on wheat sensitivities. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Rapp, M.; Schwadorf, K.; Leiser, W.L.; Würschum, T.; Longin, C.F.H. Assessing the variation and genetic architecture of asparagine content in wheat: What can plant breeding contribute to a reduction in the acrylamide precursor? Theor. Appl. Genet. 2018, 131, 2427–2437. [Google Scholar] [CrossRef]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sielaff, M.; Curella, V.; Neerukonda, M.; Afzal, M.; El Hassouni, K.; Distler, U.; Schuppan, D.; Longin, C.F.H.; Tenzer, S. Hybrid QconCAT-Based Targeted Absolute and Data-Independent Acquisition-Based Label-Free Quantification Enables In-Depth Proteomic Characterization of Wheat Amylase/Trypsin Inhibitor Extracts. J. Proteome Res. 2021. [Google Scholar] [CrossRef]
- Zetzsche, H.; Friedt, W.; Ordon, F. Breeding progress for pathogen resistance is a second major driver for yield increase in German winter wheat at contrasting N levels. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef]
- Wang, G.-F.; Wei, X.; Fan, R.; Zhou, H.; Wang, X.; Yu, C.; Dong, L.; Dong, Z.; Wang, X.; Kang, Z.; et al. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): Functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 2011, 191, 418–431. [Google Scholar] [CrossRef]
- Branlard, G.; Dardevet, M.; Amiour, N.; Igrejas, G. Allelic diversity of HMW and LMW glutenin subunits and omega-gliadins in French bread wheat (Triticum aestivum L.). Genet. Resour. Crop. Evol. 2003, 50, 669–679. [Google Scholar] [CrossRef]
- Eagles, H.A.; Cane, K.; Eastwood, R.F.; Hollamby, G.J.; Kuchel, H.; Martin, P.J.; Cornish, G.B. Contributions of glutenin and puroindoline genes to grain quality traits in southern Australian wheat breeding programs. Aust. J. Agric. Res. 2006, 57, 179–186. [Google Scholar] [CrossRef]
- Mohler, V.; Schmolke, M.; Paladey, E.; Seling, S.; Hartl, L. Association analysis of Puroindoline-D1 and Puroindoline b-2 loci with 13 quality traits in European winter wheat (Triticum aestivum L.). J. Cereal Sci. 2012, 56, 623–628. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Chang, H.-C.; Simon-Buss, A. Deciphering the immunogenic potential of wheat flour: A reference map of the salt-soluble proteome from the U.S. wheat Butte 86. Proteome Sci. 2020, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Distler, U.; Łącki, M.K.; Schumann, S.; Wanninger, M.; Tenzer, S. Enhancing Sensitivity of Microflow-Based Bottom-Up Proteomics through Postcolumn Solvent Addition. Anal. Chem. 2019, 91, 7510–7515. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.H.; Carruthers, R.; Hoyes, J.B.; Jones, C.; Langridge, J.I.; Millar, A.; Vissers, J.P.C. A novel precursor ion discovery method on a hybrid quadrupole orthogonal acceleration time-of-flight (Q-TOF) mass spectrometer for studying protein phosphorylation. J. Am. Soc. Mass Spectrom. 2002, 13, 792–803. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2013, 11, 167–170. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.-Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute Quantification of Proteins by LCMSE. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The Proteomics Identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2012, 41, D1063–D1069. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Cochran, W.G.; Cox, G.M. Experimental Designs, 2nd ed.; Wiley: New York, NY, USA, 1957. [Google Scholar]
- Stram, D.O.; Lee, J.W. Variance Components Testing in the Longitudinal Mixed Effects Model. Biometrics 1994, 50, 1171. [Google Scholar] [CrossRef] [PubMed]
- Piepho, H.-P.; Möhring, J. Computing Heritability and Selection Response from Unbalanced Plant Breeding Trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Gilmour, A.R.; Gogel, B.J.; Cullis, B.R.; Thompson, R. ASReml User Guide Release 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2009. [Google Scholar]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple comparisons among means. J. Am. Stat. Assoc. 1961, 56, 52–64. [Google Scholar] [CrossRef]

| IPN. | UniProt Accession | UniProt Annotation | InterPro Annotation | Heritability | Missing Data a (%) | PV2 b (%) | PV3 c (%) |
|---|---|---|---|---|---|---|---|
| prot034 u | A0A3B6EGL9 | Uncharacterized | Aspartic peptidase A1 family;Saposin-like type B | 0.65 | 0.21 | 100 | 99 |
| prot036 d | A0A3B6MYZ0 | Uncharacterized | Cupin 1;RmlC-like jelly roll fold | 0.72 | 0.21 | 100 | 100 |
| prot045 u | A0A3B6JER7 | Uncharacterized | Cupin 1;RmlC-like jelly roll fold | 0.83 | 0 | 100 | 100 |
| prot123 u | A0A3B6HSA4 | Uncharacterized | Cupin 1;RmlC-like jelly roll fold | 0.68 | 0 | 100 | 100 |
| prot156 d | A0A3B6LUV8 | Uncharacterized | Cupin 1;RmlC-like jelly roll fold | 0.71 | 0 | 100 | 100 |
| prot139 u | I1XB56 | Low-molecular-weight glutenin subunit | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.83 | 8.85 | 90 | 84 |
| prot245 u | D2KFG9 | Gliadin/avenin-like seed protein | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.61 | 0 | 100 | 100 |
| prot292 u | R9YQY9 | Low-molecular-weight glutenin subunit Glu-D3 | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.45 | 0.62 | 100 | 98 |
| prot069 u | A0A3B6C1C0 | Uncharacterized | Glycoside hydrolase family 1 | 0.75 | 0 | 100 | 100 |
| prot073 u | A0A1D5V0T8 | Uncharacterized | Glycoside hydrolase family 1 | 0.69 | 0.21 | 100 | 99 |
| prot018 u | F4Y589 | Heat shock protein 90 | Heat shock protein Hsp90 | 0.60 | 0.21 | 100 | 99 |
| prot218 d | P02276 | Histone H2A.2.1 | Histone-fold;Histone H2A | 0.67 | 0 | 100 | 100 |
| prot240 d | Q9SWU3 | Histone H1 WH1A.1 | Histone H1/H5 | 0.55 | 0 | 100 | 100 |
| prot222 u | A0A024CKY0 | LEA protein | Late embryogenesis abundant protein, LEA_4 subgroup | 0.34 | 2.06 | 100 | 93 |
| prot093 d | P30570 | EC protein III | Plant EC metallothionein-like protein, family 15 | 0.64 | 4.94 | 100 | 84 |
| prot068 u | A0A3B5YRZ8 | Uncharacterized | Proteinase inhibitor I13 | 0.58 | 12.76 | 91 | 74 |
| prot102 d | A0A3B6JQP1 | Uncharacterized | Cereal seed allergen/grain softness/Trypsin and alpha-amylase inhibitor | 0.25 | 0 | 100 | 100 |
| prot233 d | A0A3B6SMC2 | Uncharacterized | Ubiquitin domain;Ubiquitin-like domain superfamily | 0.13 | 0.21 | 100 | 99 |
| IPN | UniProt Accession | UniProt Annotation | InterPro Annotation | Heritability | CV (%) | Missing Data a (%) | PV2 b (%) | PV3 c (%) |
|---|---|---|---|---|---|---|---|---|
| prot085 | Q7X9M2 | Beta-amylase (Fragment) | Glycoside hydrolase, family 14 | 0.74 | 31.77 | 1.65 | 100 | 95 |
| prot141 | A0A3B6KSH4 | Beta-amylase | Glycoside hydrolase, family 14 | 0.85 | 36.89 | 0 | 100 | 100 |
| prot144 | A0A3B6IYD4 | Beta-amylase | Glycoside hydrolase, family 14 | 0.81 | 25.07 | 0 | 100 | 100 |
| prot028 | Q94G97 ⴕ | Gamma-gliadin | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.91 | 59.97 | 15.64 | 81 | 72 |
| prot066 | D2KFH0 ⴕ | Gliadin/avenin-like seed protein | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.83 | 59.31 | 19.96 | 81 | 61 |
| prot245 | D2KFG9 ⴕ | Gliadin/avenin-like seed protein | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily… Gliadin/LMW glutenin | 0.61 | 23.30 | 0 | 100 | 100 |
| prot017 | G1E6K7 ⴕ | High molecular weight glutenin subunit Dx5 (Fragment) | Bifunctional inhibitor/plant lipid transfer protein/seed storage…HMW glutenin | 0.76 | 41.79 | 1.65 | 100 | 95 |
| prot103 | Q03871 ⴕⴕ | HMW glutenin subunit 1By9 | Cereal seed allergen/grain softness/trypsin and alpha-amylase inhibitor… HMW glutenin | 0.65 | 40.63 | 6.38 | 96 | 85 |
| prot139 | I1XB56 ⴕ | Low-molecular-weight glutenin subunit (Fragment) | Bifunctional inhibitor/plant lipid transfer protein/seed storage …LMW-GS | 0.83 | 46.97 | 8.85 | 90 | 84 |
| prot203 | C3VN75 ⴕ | Low molecular weight glutenin GN = Glu-A3 | Bifunctional inhibitor/plant lipid transfer protein/seed storage …LMW-GS | 0.67 | 55.83 | 15.23 | 89 | 65 |
| prot044 | A0A3B6HWI6 | Uncharacterized | Heat shock protein 70, conserved site | 0.66 | 22.21 | 7.20 | 96 | 83 |
| prot009 | H2DLU3 ⴕ | Puroindoline b GN = Pinb-D1 | Cereal seed allergen/grain softness/trypsin and alpha-amylase inhibitor | 0.92 | 67.59 | 3.29 | 97 | 93 |
| prot110 | P81713 | Bowman-Birk type trypsin inhibitor | Bowman-Birk type proteinase inhibitor | 0.90 | 76.72 | 15.23 | 83 | 68 |
| prot030 | Q43691 ⴕ | Trypsin/alpha-amylase inhibitor CMX2 | Cereal seed allergen/grain softness/trypsin and alpha-amylase inhibitor | 0.64 | 25.12 | 12.96 | 97 | 61 |
| prot072 | Q0Q5D9 ⴕ | Globulin 1 | Cereal seed allergen/grain softness/trypsin and alpha-amylase inhibitor | 0.66 | 18.68 | 0 | 100 | 100 |
| prot288 | A0A1D5UB33 ⴕ | Uncharacterized | Cereal seed allergen/grain softness/trypsin and alpha-amylase inhibitor | 0.95 | 106.86 | 0 | 100 | 100 |
| prot189 | Q2PCC3 ⴕⴕ | Type 2 non specific lipid transfer protein | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily | 0.85 | 32.02 | 3.70 | 98 | 90 |
| prot235 | Q2PCC5 ⴕⴕ | Type 2 non specific lipid transfer protein | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily | 0.93 | 28.71 | 0 | 100 | 100 |
| prot232 | A0A3B6TLW2 | Uncharacterized | Serpin, conserved site;Serpin domain | 0.81 | 24.60 | 0.62 | 100 | 98 |
| prot287 | A0A3B6LS85 | Uncharacterized | Serpin, conserved site;Serpin domain | 0.66 | 27.39 | 9.88 | 98 | 70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afzal, M.; Sielaff, M.; Curella, V.; Neerukonda, M.; El Hassouni, K.; Schuppan, D.; Tenzer, S.; Longin, C.F.H. Characterization of 150 Wheat Cultivars by LC-MS-Based Label-Free Quantitative Proteomics Unravels Possibilities to Design Wheat Better for Baking Quality and Human Health. Plants 2021, 10, 424. https://doi.org/10.3390/plants10030424
Afzal M, Sielaff M, Curella V, Neerukonda M, El Hassouni K, Schuppan D, Tenzer S, Longin CFH. Characterization of 150 Wheat Cultivars by LC-MS-Based Label-Free Quantitative Proteomics Unravels Possibilities to Design Wheat Better for Baking Quality and Human Health. Plants. 2021; 10(3):424. https://doi.org/10.3390/plants10030424
Chicago/Turabian StyleAfzal, Muhammad, Malte Sielaff, Valentina Curella, Manjusha Neerukonda, Khaoula El Hassouni, Detlef Schuppan, Stefan Tenzer, and C. Friedrich H. Longin. 2021. "Characterization of 150 Wheat Cultivars by LC-MS-Based Label-Free Quantitative Proteomics Unravels Possibilities to Design Wheat Better for Baking Quality and Human Health" Plants 10, no. 3: 424. https://doi.org/10.3390/plants10030424
APA StyleAfzal, M., Sielaff, M., Curella, V., Neerukonda, M., El Hassouni, K., Schuppan, D., Tenzer, S., & Longin, C. F. H. (2021). Characterization of 150 Wheat Cultivars by LC-MS-Based Label-Free Quantitative Proteomics Unravels Possibilities to Design Wheat Better for Baking Quality and Human Health. Plants, 10(3), 424. https://doi.org/10.3390/plants10030424